Combinatorial optimization of cystine-knot peptides towards high-affinity inhibitors of human matriptase-1
- PMID: 24146945
- PMCID: PMC3795654
- DOI: 10.1371/journal.pone.0076956
Combinatorial optimization of cystine-knot peptides towards high-affinity inhibitors of human matriptase-1
Abstract
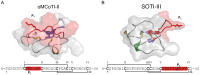
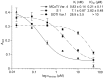
Cystine-knot miniproteins define a class of bioactive molecules with several thousand natural members. Their eponymous motif comprises a rigid structured core formed by six disulfide-connected cysteine residues, which accounts for its exceptional stability towards thermic or proteolytic degradation. Since they display a remarkable sequence tolerance within their disulfide-connected loops, these molecules are considered promising frameworks for peptide-based pharmaceuticals. Natural open-chain cystine-knot trypsin inhibitors of the MCoTI (Momordica cochinchinensis trypsin inhibitor) and SOTI (Spinacia oleracea trypsin inhibitor) families served as starting points for the generation of inhibitors of matriptase-1, a type II transmembrane serine protease with possible clinical relevance in cancer and arthritic therapy. Yeast surface-displayed libraries of miniproteins were used to select unique and potent matriptase-1 inhibitors. To this end, a knowledge-based library design was applied that makes use of detailed information on binding and folding behavior of cystine-knot peptides. Five inhibitor variants, four of the MCoTI family and one of the SOTI family, were identified, chemically synthesized and oxidatively folded towards the bioactive conformation. Enzyme assays revealed inhibition constants in the low nanomolar range for all candidates. One subnanomolar binder (Ki = 0.83 nM) with an inverted selectivity towards trypsin and matriptase-1 was identified.
Conflict of interest statement
Figures


Similar articles
-
Fragmentation follows structure: top-down mass spectrometry elucidates the topology of engineered cystine-knot miniproteins.PLoS One. 2014 Oct 10;9(10):e108626. doi: 10.1371/journal.pone.0108626. eCollection 2014. PLoS One. 2014. PMID: 25303319 Free PMC article.
-
Chemical synthesis, backbone cyclization and oxidative folding of cystine-knot peptides: promising scaffolds for applications in drug design.Molecules. 2012 Oct 24;17(11):12533-52. doi: 10.3390/molecules171112533. Molecules. 2012. PMID: 23095896 Free PMC article. Review.
-
Computational analysis of the MCoTI-II plant defence knottin reveals a novel intermediate conformation that facilitates trypsin binding.Sci Rep. 2016 Mar 15;6:23174. doi: 10.1038/srep23174. Sci Rep. 2016. PMID: 26975976 Free PMC article.
-
High-affinity cyclic peptide matriptase inhibitors.J Biol Chem. 2013 May 10;288(19):13885-96. doi: 10.1074/jbc.M113.460030. Epub 2013 Apr 2. J Biol Chem. 2013. PMID: 23548907 Free PMC article.
-
Synthetic Cystine-Knot Miniproteins - Valuable Scaffolds for Polypeptide Engineering.Adv Exp Med Biol. 2016;917:121-44. doi: 10.1007/978-3-319-32805-8_7. Adv Exp Med Biol. 2016. PMID: 27236555 Review.
Cited by
-
High Proteolytic Resistance of Spider-Derived Inhibitor Cystine Knots.Int J Pept. 2015;2015:537508. doi: 10.1155/2015/537508. Epub 2015 Dec 30. Int J Pept. 2015. PMID: 26843868 Free PMC article.
-
Ultra-High-Throughput Screening of Cystine-Rich Peptide Libraries Via Yeast Surface Display.Methods Mol Biol. 2025;2934:275-291. doi: 10.1007/978-1-0716-4578-9_18. Methods Mol Biol. 2025. PMID: 40663336
-
Applications of Yeast Surface Display for Protein Engineering.Methods Mol Biol. 2015;1319:155-75. doi: 10.1007/978-1-4939-2748-7_8. Methods Mol Biol. 2015. PMID: 26060074 Free PMC article. Review.
-
Mammalian display screening of diverse cystine-dense peptides for difficult to drug targets.Nat Commun. 2017 Dec 21;8(1):2244. doi: 10.1038/s41467-017-02098-8. Nat Commun. 2017. PMID: 29269835 Free PMC article.
-
Plant peptides - redefining an area of ribosomally synthesized and post-translationally modified peptides.Nat Prod Rep. 2024 Jul 17;41(7):1020-1059. doi: 10.1039/d3np00042g. Nat Prod Rep. 2024. PMID: 38411572 Free PMC article. Review.
References
-
- Chiche L, Heitz A, Gelly JC, Gracy J, Chau PT, et al. (2004) Squash inhibitors: from structural motifs to macrocyclic knottins. Curr Protein Pept Sci 5: 341–349. - PubMed
-
- Kolmar H (2009) Biological diversity and therapeutic potential of natural and engineered cystine knot miniproteins. Curr Opin Pharmacol 9: 608–614. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

