Comparison of the structural and dynamic effects of 5-methylcytosine and 5-chlorocytosine in a CpG dinucleotide sequence
- PMID: 24147911
- PMCID: PMC3985228
- DOI: 10.1021/bi400980c
Comparison of the structural and dynamic effects of 5-methylcytosine and 5-chlorocytosine in a CpG dinucleotide sequence
Abstract
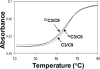
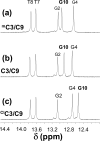
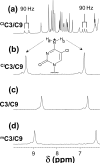
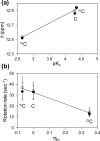
Inflammation-mediated reactive molecules can result in an array of oxidized and halogenated DNA-damage products, including 5-chlorocytosine ((Cl)C). Previous studies have shown that (Cl)C can mimic 5-methylcytosine ((m)C) and act as a fraudulent epigenetic signal, promoting the methylation of previously unmethylated DNA sequences. Although the 5-halouracils are good substrates for base-excision repair, no repair activity has yet been identified for (Cl)C. Because of the apparent biochemical similarities of (m)C and (Cl)C, we have investigated the effects of (m)C and (Cl)C substitution on oligonucleotide structure and dynamics. In this study, we have constructed oligonucleotide duplexes containing C, (Cl)C, and (m)C within a CpG dinucleotide. The thermal and thermodynamic stability of these duplexes were found to be experimentally indistinguishable. Crystallographic structures of duplex oligonucleotides containing (m)C or (Cl)C were determined to 1.2 and 1.9 Å resolution, respectively. Both duplexes are B-form and are superimposable on a previously determined structure of a cytosine-containing duplex with a rmsd of approximately 0.25 Å. NMR solution studies indicate that all duplexes containing cytosine or the cytosine analogues are normal B-form and that no structural perturbations are observed surrounding the site of each substitution. The magnitude of the base-stacking-induced upfield shifts for nonexchangeable base proton resonances are similar for each of the duplexes examined, indicating that neither (m)C nor (Cl)C significantly alter base-stacking interactions. The (Cl)C analogue is paired with G in an apparently normal geometry; however, the G-imino proton of the (Cl)C-G base pair resonates to higher field relative to (m)C-G or C-G, indicating a weaker imino hydrogen bond. Using selective ¹⁵N-enrichment and isotope-edited NMR, we observe that the amino group of (Cl)C rotates at roughly half of the rate of the corresponding amino groups of the C-G and (m)C-G base pairs. The altered chemical shifts of hydrogen-bonding proton resonances for the (Cl)C-G base pair as well as the slower rotation of the (Cl)C amino group can be attributed to the electron-withdrawing inductive property of the 5-chloro substituent. The apparent similarity of duplexes containing (m)C and (Cl)C demonstrated here is in accord with results of previous biochemical studies and further suggests that (Cl)C is likely to be an unusually persistent form of DNA damage.
Figures



Similar articles
-
Direct evidence for (G)O6···H2-N4(C)+ hydrogen bonding in transient G(syn)-C+ and G(syn)-m5C+ Hoogsteen base pairs in duplex DNA from cytosine amino nitrogen off-resonance R1ρ relaxation dispersion measurements.J Magn Reson. 2019 Nov;308:106589. doi: 10.1016/j.jmr.2019.106589. Epub 2019 Sep 5. J Magn Reson. 2019. PMID: 31539864 Free PMC article.
-
Thermodynamic stability and solution conformation of tandem G.A mismatches in RNA and RNA.DNA hybrid duplexes.Eur J Biochem. 1994 Mar 15;220(3):703-15. doi: 10.1111/j.1432-1033.1994.tb18671.x. Eur J Biochem. 1994. PMID: 8143725
-
Sequence dependent effects of CpG cytosine methylation. A joint 1H-NMR and 31P-NMR study.Eur J Biochem. 1995 Apr 15;229(2):445-54. doi: 10.1111/j.1432-1033.1995.0445k.x. Eur J Biochem. 1995. PMID: 7744067
-
Effect of environment, conformation, sequence and base substituents on the imino proton exchange rates in guanine and inosine-containing DNA, RNA, and DNA-RNA duplexes.J Mol Biol. 1984 Aug 5;177(2):207-27. doi: 10.1016/0022-2836(84)90453-4. J Mol Biol. 1984. PMID: 6205159
-
Epigenetic Modifications of Cytosine: Biophysical Properties, Regulation, and Function in Mammalian DNA.Bioessays. 2018 Mar;40(3). doi: 10.1002/bies.201700199. Epub 2018 Jan 25. Bioessays. 2018. PMID: 29369386 Review.
Cited by
-
Intrinsic mutagenic properties of 5-chlorocytosine: A mechanistic connection between chronic inflammation and cancer.Proc Natl Acad Sci U S A. 2015 Aug 18;112(33):E4571-80. doi: 10.1073/pnas.1507709112. Epub 2015 Aug 4. Proc Natl Acad Sci U S A. 2015. PMID: 26243878 Free PMC article.
-
Combined Effects of Methylated Cytosine and Molecular Crowding on the Thermodynamic Stability of DNA Duplexes.Int J Mol Sci. 2021 Jan 19;22(2):947. doi: 10.3390/ijms22020947. Int J Mol Sci. 2021. PMID: 33477917 Free PMC article.
-
Structural basis of water-mediated cis Watson-Crick/Hoogsteen base-pair formation in non-CpG methylation.Nucleic Acids Res. 2024 Aug 12;52(14):8566-8579. doi: 10.1093/nar/gkae594. Nucleic Acids Res. 2024. PMID: 38989613 Free PMC article.
-
Synthesis and DNA/RNA complementation studies of peptide nucleic acids containing 5-halouracils.Medchemcomm. 2016 Dec 12;8(2):385-389. doi: 10.1039/c6md00536e. eCollection 2017 Feb 1. Medchemcomm. 2016. PMID: 30108754 Free PMC article.
-
Cytosine methylation of mitochondrial DNA at CpG sequences impacts transcription factor A DNA binding and transcription.Biochim Biophys Acta Gene Regul Mech. 2019 May;1862(5):598-607. doi: 10.1016/j.bbagrm.2019.01.006. Epub 2019 Feb 23. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 30807854 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

