Comparative analysis of resistant and susceptible macrophage gene expression response to Leishmania major parasite
- PMID: 24148319
- PMCID: PMC4007596
- DOI: 10.1186/1471-2164-14-723
Comparative analysis of resistant and susceptible macrophage gene expression response to Leishmania major parasite
Abstract
Background: Leishmania are obligated intracellular pathogens that replicate almost exclusively in macrophages. The outcome of infection depends largely on parasite pathogenicity and virulence but also on the activation status and genetic background of macrophages. Animal models are essential for a better understanding of pathogenesis of different microbes including Leishmania.
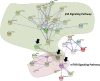
Results: Here we compared the transcriptional signatures of resistant (C57BL/6) and susceptible (BALB/c) mouse bone marrow-derived macrophages in response to Leishmania major (L. major) promastigotes infection.Microarray results were first analyzed for significant pathways using the Kyoto Encylopedia of Genes and Genomes (KEGG) database. The analysis revealed that a large set of the shared genes is involved in the immune response and that difference in the expression level of some chemokines and chemokine receptors could partially explain differences in resistance. We next focused on up-regulated genes unique to either BALB/c or C57BL/6 derived macrophages and identified, using KEGG database, signal transduction pathways among the most relevant pathways unique to both susceptible and resistant derived macrophages. Indeed, genes unique to C57BL/6 BMdMs were associated with target of rapamycin (mTOR) signaling pathway while a range of genes unique to BALB/c BMdMs, belong to p53 signaling pathway. We next investigated whether, in a given mice strain derived macrophages, the different up-regulated unique genes could be coordinately regulated. Using GeneMapp Cytoscape, we showed that the induced genes unique to BALB/c or C57BL/6 BMdMs are interconnected. Finally, we examined whether the induced pathways unique to BALB/c derived macrophages interfere with the ones unique to C57BL/6 derived macrophages. Protein-protein interaction analysis using String database highlights the existence of a cross-talk between p53 and mTOR signaling pathways respectively specific to susceptible and resistant BMdMs.
Conclusions: Taken together our results suggest that strains specific pathogenesis may be due to a difference in the magnitude of the same pathways and/or to differentially expressed pathways in the two mouse strains derived macrophages. We identify signal transduction pathways among the most relevant pathways modulated by L. major infection, unique to BALB/c and C57BL/6 BMdM and postulate that the interplay between these potentially interconnected pathways could direct the macrophage response toward a given phenotype.
Figures



References
-
- Buates S, Matlashewski G. General suppression of macrophage gene expression during Leishmania donovani infection. J Immunol. 2001;14(5):3416–3422. - PubMed
-
- Guerfali FZ, Laouini D, Guizani-Tabbane L, Ottones F, Ben-Aissa K, Benkahla A, Manchon L, Piquemal D, Smandi S, Mghirbi O. et al.Simultaneous gene expression profiling in human macrophages infected with Leishmania major parasites using SAGE. BMC Genomics. 2008;14:238. doi: 10.1186/1471-2164-9-238. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

