DNA repair in Mycoplasma gallisepticum
- PMID: 24148612
- PMCID: PMC4007778
- DOI: 10.1186/1471-2164-14-726
DNA repair in Mycoplasma gallisepticum
Abstract
Background: DNA repair is essential for the maintenance of genome stability in all living beings. Genome size as well as the repertoire and abundance of DNA repair components may vary among prokaryotic species. The bacteria of the Mollicutes class feature a small genome size, absence of a cell wall, and a parasitic lifestyle. A small number of genes make Mollicutes a good model for a "minimal cell" concept.
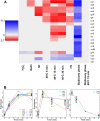
Results: In this work we studied the DNA repair system of Mycoplasma gallisepticum on genomic, transcriptional, and proteomic levels. We detected 18 out of 22 members of the DNA repair system on a protein level. We found that abundance of the respective mRNAs is less than one per cell. We studied transcriptional response of DNA repair genes of M. gallisepticum at stress conditions including heat, osmotic, peroxide stresses, tetracycline and ciprofloxacin treatment, stationary phase and heat stress in stationary phase.
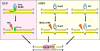
Conclusions: Based on comparative genomic study, we determined that the DNA repair system M. gallisepticum includes a sufficient set of proteins to provide a cell with functional nucleotide and base excision repair and mismatch repair. We identified SOS-response in M. gallisepticum on ciprofloxacin, which is a known SOS-inducer, tetracycline and heat stress in the absence of established regulators. Heat stress was found to be the strongest SOS-inducer. We found that upon transition to stationary phase of culture growth transcription of DNA repair genes decreases dramatically. Heat stress does not induce SOS-response in a stationary phase.
Figures

References
-
- Alexeev D, Kostrjukova E, Aliper A, Popenko A, Bazaleev N, Tyakht A, Selezneva O, Akopian T, Prichodko E, Kondratov I, Chukin M, Demina I, Galyamina M, Kamashev D, Vanyushkina A, Ladygina V, Levitskii S, Lazarev V, Govorun V. Application of Spiroplasma melliferum proteogenomic profiling for the discovery of virulence factors and pathogenicity mechanisms in host-associated spiroplasmas. J Proteome Res. 2012;14:224–236. doi: 10.1021/pr2008626. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

