Perspectives on the mechanism of transcriptional regulation by long non-coding RNAs
- PMID: 24149621
- PMCID: PMC3928176
- DOI: 10.4161/epi.26700
Perspectives on the mechanism of transcriptional regulation by long non-coding RNAs
Abstract
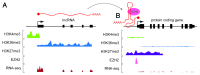
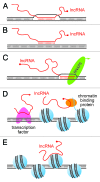
Long non-coding RNAs (lncRNAs) are increasingly being recognized as epigenetic regulators of gene transcription. The diversity and complexity of lncRNA genes means that they exert their regulatory effects by a variety of mechanisms. Although there is still much to be learned about the mechanism of lncRNA function, general principles are starting to emerge. In particular, the application of high throughput (deep) sequencing methodologies has greatly advanced our understanding of lncRNA gene function. lncRNAs function as adaptors that link specific chromatin loci with chromatin-remodeling complexes and transcription factors. lncRNAs can act in cis or trans to guide epigenetic-modifier complexes to distinct genomic sites, or act as scaffolds which recruit multiple proteins simultaneously, thereby coordinating their activities. In this review we discuss the genomic organization of lncRNAs, the importance of RNA secondary structure to lncRNA functionality, the multitude of ways in which they interact with the genome, and what evolutionary conservation tells us about their function.
Keywords: RNA; RNA scaffolds; RNA structure; RNAi; TGA; TGS; chromatin; epigenetics; lncRNA; ncRNA.
Figures
Similar articles
-
Long non-coding RNA and chromatin remodeling.RNA Biol. 2015;12(10):1094-8. doi: 10.1080/15476286.2015.1063770. Epub 2015 Jul 15. RNA Biol. 2015. PMID: 26177256 Free PMC article. Review.
-
Epigenetic regulation by long noncoding RNAs in plants.Chromosome Res. 2013 Dec;21(6-7):685-93. doi: 10.1007/s10577-013-9392-6. Chromosome Res. 2013. PMID: 24233054 Free PMC article. Review.
-
Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer.Genomics Proteomics Bioinformatics. 2016 Feb;14(1):42-54. doi: 10.1016/j.gpb.2015.09.006. Epub 2016 Feb 12. Genomics Proteomics Bioinformatics. 2016. PMID: 26883671 Free PMC article. Review.
-
Long noncoding RNA in genome regulation: prospects and mechanisms.RNA Biol. 2010 Sep-Oct;7(5):582-5. doi: 10.4161/rna.7.5.13216. Epub 2010 Sep 1. RNA Biol. 2010. PMID: 20930520 Free PMC article.
-
R-Loop Mediated trans Action of the APOLO Long Noncoding RNA.Mol Cell. 2020 Mar 5;77(5):1055-1065.e4. doi: 10.1016/j.molcel.2019.12.015. Epub 2020 Jan 14. Mol Cell. 2020. PMID: 31952990
Cited by
-
TCF-4 Regulated lncRNA-XIST Promotes M2 Polarization Of Macrophages And Is Associated With Lung Cancer.Onco Targets Ther. 2019 Oct 2;12:8055-8062. doi: 10.2147/OTT.S210952. eCollection 2019. Onco Targets Ther. 2019. PMID: 31632059 Free PMC article.
-
Uncovering missed indels by leveraging unmapped reads.Sci Rep. 2019 Jul 31;9(1):11093. doi: 10.1038/s41598-019-47405-z. Sci Rep. 2019. PMID: 31366961 Free PMC article.
-
Genome‑wide identification of long noncoding RNAs in CCl4‑induced liver fibrosis via RNA sequencing.Mol Med Rep. 2018 Jul;18(1):299-307. doi: 10.3892/mmr.2018.8986. Epub 2018 May 7. Mol Med Rep. 2018. PMID: 29749545 Free PMC article.
-
Correlation of long non-coding RNA expression with metastasis, drug resistance and clinical outcome in cancer.Oncotarget. 2014 Sep 30;5(18):8027-38. doi: 10.18632/oncotarget.2469. Oncotarget. 2014. PMID: 25275300 Free PMC article. Review.
-
Downregulation of long noncoding RNA NONHSAT037832 in papillary thyroid carcinoma and its clinical significance.Tumour Biol. 2016 May;37(5):6117-23. doi: 10.1007/s13277-015-4461-4. Epub 2015 Nov 26. Tumour Biol. 2016. PMID: 26611646
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
