Overlapping and distinct functions of CstF64 and CstF64τ in mammalian mRNA 3' processing
- PMID: 24149845
- PMCID: PMC3884657
- DOI: 10.1261/rna.042317.113
Overlapping and distinct functions of CstF64 and CstF64τ in mammalian mRNA 3' processing
Abstract
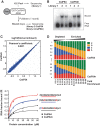
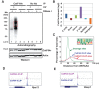
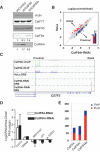
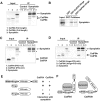
mRNA 3' processing is dynamically regulated spatially and temporally. However, the underlying mechanisms remain poorly understood. CstF64τ is a paralog of the general mRNA 3' processing factor, CstF64, and has been implicated in mediating testis-specific mRNA alternative polyadenylation (APA). However, the functions of CstF64τ in mRNA 3' processing have not been systematically investigated. We carried out a comprehensive characterization of CstF64τ and compared its properties to those of CstF64. In contrast to previous reports, we found that both CstF64 and CstF64τ are widely expressed in mammalian tissues, and their protein levels display tissue-specific variations. We further demonstrated that CstF64 and CstF64τ have highly similar RNA-binding specificities both in vitro and in vivo. CstF64 and CstF64τ modulate one another's expression and play overlapping as well as distinct roles in regulating global APA profiles. Interestingly, protein interactome analyses revealed key differences between CstF64 and CstF64τ, including their interactions with another mRNA 3' processing factor, symplekin. Together, our study of CstF64 and CstF64τ revealed both functional overlap and specificity of these two important mRNA 3' processing factors and provided new insights into the regulatory mechanisms of mRNA 3' processing.
Keywords: 3′ end formation; alternative polyadenylation; mRNA processing; sequencing.
Figures


Similar articles
-
Transcriptome-wide analyses of CstF64-RNA interactions in global regulation of mRNA alternative polyadenylation.Proc Natl Acad Sci U S A. 2012 Nov 13;109(46):18773-8. doi: 10.1073/pnas.1211101109. Epub 2012 Oct 29. Proc Natl Acad Sci U S A. 2012. PMID: 23112178 Free PMC article.
-
CstF64: cell cycle regulation and functional role in 3' end processing of replication-dependent histone mRNAs.Mol Cell Biol. 2014 Dec 1;34(23):4272-84. doi: 10.1128/MCB.00791-14. Epub 2014 Sep 29. Mol Cell Biol. 2014. PMID: 25266659 Free PMC article.
-
3'-End processing of histone pre-mRNAs in Drosophila: U7 snRNP is associated with FLASH and polyadenylation factors.RNA. 2013 Dec;19(12):1726-44. doi: 10.1261/rna.040360.113. Epub 2013 Oct 21. RNA. 2013. PMID: 24145821 Free PMC article.
-
Alternative polyadenylation of mRNA precursors.Nat Rev Mol Cell Biol. 2017 Jan;18(1):18-30. doi: 10.1038/nrm.2016.116. Epub 2016 Sep 28. Nat Rev Mol Cell Biol. 2017. PMID: 27677860 Free PMC article. Review.
-
Implications of polyadenylation in health and disease.Nucleus. 2014;5(6):508-19. doi: 10.4161/nucl.36360. Epub 2014 Oct 31. Nucleus. 2014. PMID: 25484187 Free PMC article. Review.
Cited by
-
APC mutations dysregulate alternative polyadenylation in cancer.Genome Biol. 2024 Oct 7;25(1):255. doi: 10.1186/s13059-024-03406-4. Genome Biol. 2024. PMID: 39375704 Free PMC article.
-
Identifying human pre-mRNA cleavage and polyadenylation factors by genome-wide CRISPR screens using a dual fluorescence readthrough reporter.Nucleic Acids Res. 2024 May 8;52(8):4483-4501. doi: 10.1093/nar/gkae240. Nucleic Acids Res. 2024. PMID: 38587191 Free PMC article.
-
The end of the message: multiple protein-RNA interactions define the mRNA polyadenylation site.Genes Dev. 2015 May 1;29(9):889-97. doi: 10.1101/gad.261974.115. Genes Dev. 2015. PMID: 25934501 Free PMC article. Review.
-
Cleavage stimulating factor 64 depletion mitigates cardiac fibrosis through alternative polyadenylation.Biochem Biophys Res Commun. 2022 Mar 15;597:109-114. doi: 10.1016/j.bbrc.2022.01.093. Epub 2022 Jan 29. Biochem Biophys Res Commun. 2022. PMID: 35134608 Free PMC article.
-
The polyadenylation code: a unified model for the regulation of mRNA alternative polyadenylation.J Zhejiang Univ Sci B. 2014 May;15(5):429-37. doi: 10.1631/jzus.B1400076. J Zhejiang Univ Sci B. 2014. PMID: 24793760 Free PMC article. Review.
References
-
- Bai Y, Auperin TC, Chou CY, Chang GG, Manley JL, Tong L. 2007. Crystal structure of murine CstF-77: Dimeric association and implications for polyadenylation of mRNA precursors. Mol Cell 25: 863–875. - PubMed
-
- Colgan DF, Manley JL. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev 11: 2755–2766. - PubMed
-
- Coutinho-Mansfield GC, Xue Y, Zhang Y, Fu XD. 2007. PTB/nPTB switch: A post-transcriptional mechanism for programming neuronal differentiation. Genes Dev 21: 1573–1577. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- P41 GM103533/GM/NIGMS NIH HHS/United States
- R01 GM088342/GM/NIGMS NIH HHS/United States
- R01 GM090056/GM/NIGMS NIH HHS/United States
- 5P41RR011823-17/RR/NCRR NIH HHS/United States
- R01GM090056/GM/NIGMS NIH HHS/United States
- P41 RR011823/RR/NCRR NIH HHS/United States
- R01 AG027463/AG/NIA NIH HHS/United States
- R01 HG006870/HG/NHGRI NIH HHS/United States
- CDP 12-186/HX/HSRD VA/United States
- 8 P41 GM103533-17/GM/NIGMS NIH HHS/United States
- R01AG027463-04/AG/NIA NIH HHS/United States
- R01GM088342/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
