CASP10 results compared to those of previous CASP experiments
- PMID: 24150928
- PMCID: PMC4180100
- DOI: 10.1002/prot.24448
CASP10 results compared to those of previous CASP experiments
Abstract
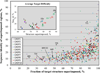
We compare results of the community efforts in modeling protein structures in the tenth CASP experiment, with those in earlier CASPs particularly in CASP5, a decade ago. There is a substantial improvement in template based model accuracy as reflected in more successful modeling of regions of structure not easily derived from a single experimental structure template, most likely reflecting intensive work within the modeling community in developing methods that make use of multiple templates, as well as the increased number of experimental structures available. Deriving structural information not obvious from a template is the most demanding as well as one of the most useful tasks that modeling can perform. Thus this is gratifying progress. By contrast, overall backbone accuracy of models appears little changed in the last decade. This puzzling result is explained by two factors--increased database size in some ways makes it harder to choose the best available templates, and the increased intrinsic difficulty of CASP targets as experimental work has progressed to larger and more unusual structures. There is no detectable recent improvement in template-free modeling, but again, this may reflect the changing nature of CASP targets.
Keywords: CASP; community wide experiment; protein structure prediction.
Copyright © 2013 Wiley Periodicals, Inc.
Figures








References
-
- Moult J. A decade of CASP: progress, bottlenecks and prognosis in protein structure prediction. Curr Opin Struct Biol. 2005;15:285–289. - PubMed
-
- Kryshtafovych A, Venclovas C, Fidelis K, Moult J. Progress over the first decade of CASP experiments. Proteins. 2005;61(Suppl 7):225–236. - PubMed
-
- Kryshtafovych A, Fidelis K, Moult J. Progress from CASP6 to CASP7. Proteins. 2007;69:194–207. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

