DECIPHER: database for the interpretation of phenotype-linked plausibly pathogenic sequence and copy-number variation
- PMID: 24150940
- PMCID: PMC3965078
- DOI: 10.1093/nar/gkt937
DECIPHER: database for the interpretation of phenotype-linked plausibly pathogenic sequence and copy-number variation
Abstract
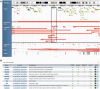
The DECIPHER database (https://decipher.sanger.ac.uk/) is an accessible online repository of genetic variation with associated phenotypes that facilitates the identification and interpretation of pathogenic genetic variation in patients with rare disorders. Contributing to DECIPHER is an international consortium of >200 academic clinical centres of genetic medicine and ≥1600 clinical geneticists and diagnostic laboratory scientists. Information integrated from a variety of bioinformatics resources, coupled with visualization tools, provides a comprehensive set of tools to identify other patients with similar genotype-phenotype characteristics and highlights potentially pathogenic genes. In a significant development, we have extended DECIPHER from a database of just copy-number variants to allow upload, annotation and analysis of sequence variants such as single nucleotide variants (SNVs) and InDels. Other notable developments in DECIPHER include a purpose-built, customizable and interactive genome browser to aid combined visualization and interpretation of sequence and copy-number variation against informative datasets of pathogenic and population variation. We have also introduced several new features to our deposition and analysis interface. This article provides an update to the DECIPHER database, an earlier instance of which has been described elsewhere [Swaminathan et al. (2012) DECIPHER: web-based, community resource for clinical interpretation of rare variants in developmental disorders. Hum. Mol. Genet., 21, R37-R44].
Figures


References
-
- Roizen NJ, Patterson D. Down's syndrome. Lancet. 2003;361:1281–1289. - PubMed
-
- Shaw-Smith C, Redon R, Rickman L, Rio M, Willatt L, Fiegler H, Firth H, Sanlaville D, Winter R, Colleaux L, et al. Microarray based comparative genomic hybridisation (array-CGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmorphic features. J. Med. Genet. 2004;41:241–248. - PMC - PubMed
-
- Stankiewicz P, Beaudet AL. Use of array CGH in the evaluation of dysmorphology, malformations, developmental delay, and idiopathic mental retardation. Curr. Opin. Genet. Dev. 2007;17:182–192. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

