Phosphatidylinositol phosphate 5-kinase Iγi2 in association with Src controls anchorage-independent growth of tumor cells
- PMID: 24151076
- PMCID: PMC3843082
- DOI: 10.1074/jbc.M113.512848
Phosphatidylinositol phosphate 5-kinase Iγi2 in association with Src controls anchorage-independent growth of tumor cells
Abstract
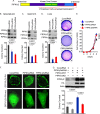
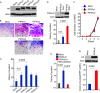
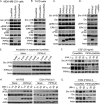
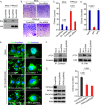
A fundamental property of tumor cells is to defy anoikis, cell death caused by a lack of cell-matrix interaction, and grow in an anchorage-independent manner. How tumor cells organize signaling molecules at the plasma membrane to sustain oncogenic signals in the absence of cell-matrix interactions remains poorly understood. Here, we describe a role for phosphatidylinositol 4-phosphate 5-kinase (PIPK) Iγi2 in controlling anchorage-independent growth of tumor cells in coordination with the proto-oncogene Src. PIPKIγi2 regulated Src activation downstream of growth factor receptors and integrins. PIPKIγi2 directly interacted with the C-terminal tail of Src and regulated its subcellular localization in concert with talin, a cytoskeletal protein targeted to focal adhesions. Co-expression of PIPKIγi2 and Src synergistically induced the anchorage-independent growth of nonmalignant cells. This study uncovers a novel mechanism where a phosphoinositide-synthesizing enzyme, PIPKIγi2, functions with the proto-oncogene Src, to regulate oncogenic signaling.
Keywords: Anchorage-independent Growth; Cancer Biology; Cell Growth; Oncogene; Phosphatidylinositol; Src; Talin.
Figures
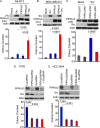
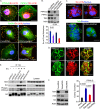
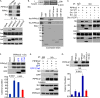
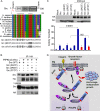
References
-
- Simpson C. D., Anyiwe K., Schimmer A. D. (2008) Anoikis resistance and tumor metastasis. Cancer Lett. 272, 177–185 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

