A mediator of singlet oxygen responses in Chlamydomonas reinhardtii and Arabidopsis identified by a luciferase-based genetic screen in algal cells
- PMID: 24151292
- PMCID: PMC3877789
- DOI: 10.1105/tpc.113.117390
A mediator of singlet oxygen responses in Chlamydomonas reinhardtii and Arabidopsis identified by a luciferase-based genetic screen in algal cells
Abstract
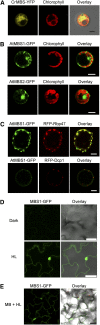
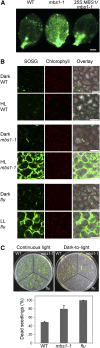
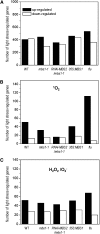
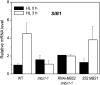
All cells produce reactive oxygen species (ROS) as by-products of their metabolism. In addition to being cytotoxic, ROS act as regulators of a wide range of developmental and physiological processes. Little is known about the molecular mechanisms underlying the perception of ROS and initiation of cellular responses in eukaryotes. Using the unicellular green alga Chlamydomonas reinhardtii, we developed a genetic screen for early components of singlet oxygen signaling. Here, we report the identification of a small zinc finger protein, methylene blue sensitivity (MBS), that is required for induction of singlet oxygen-dependent gene expression and, upon oxidative stress, accumulates in distinct granules in the cytosol. Loss-of-function mbs mutants produce singlet oxygen but are unable to fully respond to it at the level of gene expression. Knockout or knockdown of the homologous genes in the higher plant model Arabidopsis thaliana results in mutants that are hypersensitive to photooxidative stress, whereas overexpression produces plants with elevated stress tolerance. Together, our data indicate an important and evolutionarily conserved role of the MBS protein in ROS signaling and provide a strategy for engineering stress-tolerant plants.
Figures




Similar articles
-
Characterization of singlet oxygen-accumulating mutants isolated in a screen for altered oxidative stress response in Chlamydomonas reinhardtii.BMC Plant Biol. 2010 Dec 17;10:279. doi: 10.1186/1471-2229-10-279. BMC Plant Biol. 2010. PMID: 21167020 Free PMC article.
-
Superoxide-responsive gene expression in Arabidopsis thaliana and Zea mays.Plant Physiol Biochem. 2017 Aug;117:51-60. doi: 10.1016/j.plaphy.2017.05.018. Epub 2017 May 29. Plant Physiol Biochem. 2017. PMID: 28587993
-
SINGLET OXYGEN RESISTANT 1 links reactive electrophile signaling to singlet oxygen acclimation in Chlamydomonas reinhardtii.Proc Natl Acad Sci U S A. 2012 May 15;109(20):E1302-11. doi: 10.1073/pnas.1116843109. Epub 2012 Apr 23. Proc Natl Acad Sci U S A. 2012. PMID: 22529359 Free PMC article.
-
Respiratory-deficient mutants of the unicellular green alga Chlamydomonas: a review.Biochimie. 2014 May;100:207-18. doi: 10.1016/j.biochi.2013.10.006. Epub 2013 Oct 15. Biochimie. 2014. PMID: 24139906 Review.
-
Light stress and photoprotection in Chlamydomonas reinhardtii.Plant J. 2015 May;82(3):449-465. doi: 10.1111/tpj.12825. Epub 2015 Apr 9. Plant J. 2015. PMID: 25758978 Review.
Cited by
-
Oxygen and ROS in Photosynthesis.Plants (Basel). 2020 Jan 10;9(1):91. doi: 10.3390/plants9010091. Plants (Basel). 2020. PMID: 31936893 Free PMC article. Review.
-
Transcriptomic and Physiological Responses to Oxidative Stress in a Chlamydomonas reinhardtii Glutathione Peroxidase Mutant.Genes (Basel). 2020 Apr 24;11(4):463. doi: 10.3390/genes11040463. Genes (Basel). 2020. PMID: 32344528 Free PMC article.
-
FtsH2-Dependent Proteolysis of EXECUTER1 Is Essential in Mediating Singlet Oxygen-Triggered Retrograde Signaling in Arabidopsis thaliana.Front Plant Sci. 2017 Jun 29;8:1145. doi: 10.3389/fpls.2017.01145. eCollection 2017. Front Plant Sci. 2017. PMID: 28706530 Free PMC article.
-
Interaction between photosynthetic electron transport and chloroplast sinks triggers protection and signalling important for plant productivity.Philos Trans R Soc Lond B Biol Sci. 2017 Sep 26;372(1730):20160390. doi: 10.1098/rstb.2016.0390. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28808104 Free PMC article.
-
The Chlamydomonas bZIP transcription factor BLZ8 confers oxidative stress tolerance by inducing the carbon-concentrating mechanism.Plant Cell. 2022 Feb 3;34(2):910-926. doi: 10.1093/plcell/koab293. Plant Cell. 2022. PMID: 34893905 Free PMC article.
References
-
- Antoni R., Rodriguez L., Gonzalez-Guzman M., Pizzio G.A., Rodriguez P.L. (2011). News on ABA transport, protein degradation, and ABFs/WRKYs in ABA signaling. Curr. Opin. Plant Biol. 14: 547–553 - PubMed
-
- Apel K., Hirt H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55: 373–399 - PubMed
-
- Baruah A., Simková K., Apel K., Laloi C. (2009). Arabidopsis mutants reveal multiple singlet oxygen signaling pathways involved in stress response and development. Plant Mol. Biol. 70: 547–563 - PubMed
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

