A comprehensive mutational analysis of the Arabidopsis resistance protein RPW8.2 reveals key amino acids for defense activation and protein targeting
- PMID: 24151293
- PMCID: PMC3877822
- DOI: 10.1105/tpc.113.117226
A comprehensive mutational analysis of the Arabidopsis resistance protein RPW8.2 reveals key amino acids for defense activation and protein targeting
Abstract
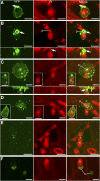
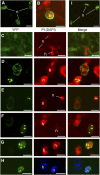
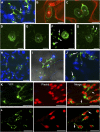
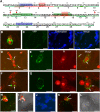
The Arabidopsis thaliana resistance to powdery mildew8.2 (RPW8.2) protein is specifically targeted to the extrahaustorial membrane (EHM) encasing the haustorium, or fungal feeding structure, where RPW8.2 activates broad-spectrum resistance against powdery mildew pathogens. How RPW8.2 activates defenses at a precise subcellular locale is not known. Here, we report a comprehensive mutational analysis in which more than 100 RPW8.2 mutants were functionally evaluated for their defense and trafficking properties. We show that three amino acid residues (i.e., threonine-64, valine-68, and aspartic acid-116) are critical for RPW8.2-mediated cell death and resistance to powdery mildew (Golovinomyces cichoracearum UCSC1). Also, we reveal that two arginine (R)- or lysine (K)-enriched short motifs (i.e., R/K-R/K-x-R/K) make up the likely core EHM-targeting signals, which, together with the N-terminal transmembrane domain, define a minimal sequence of 60 amino acids that is necessary and sufficient for EHM localization. In addition, some RPW8.2 mutants localize to the nucleus and/or to a potentially novel membrane that wraps around plastids or plastid-derived stromules. Results from this study not only reveal critical amino acid elements in RPW8.2 that enable haustorium-targeted trafficking and defense, but also provide evidence for the existence of a specific, EHM-oriented membrane trafficking pathway in leaf epidermal cells invaded by powdery mildew.
Figures



Comment in
-
Border patrol on the extrahaustorial membrane: Arabidopsis resistance protein RPW8.2 activates targeted, postpenetration defenses.Plant Cell. 2013 Oct;25(10):3638. doi: 10.1105/tpc.113.251014. Epub 2013 Oct 25. Plant Cell. 2013. PMID: 24163314 Free PMC article. No abstract available.
References
-
- Assaad F.F., Qiu J.L., Youngs H., Ehrhardt D., Zimmerli L., Kalde M., Wanner G., Peck S.C., Edwards H., Ramonell K., Somerville C.R., Thordal-Christensen H. (2004). The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15: 5118–5129 - PMC - PubMed
-
- Bhattacharjee S., Halane M.K., Kim S.H., Gassmann W. (2011). Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 334: 1405–1408 - PubMed
-
- Bonfante P., Genre A. (2010). Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nat. Commun. 1: 48. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

