In vitro binding of Sorghum bicolor transcription factors ABI4 and ABI5 to a conserved region of a GA 2-OXIDASE promoter: possible role of this interaction in the expression of seed dormancy
- PMID: 24151305
- PMCID: PMC3871824
- DOI: 10.1093/jxb/ert347
In vitro binding of Sorghum bicolor transcription factors ABI4 and ABI5 to a conserved region of a GA 2-OXIDASE promoter: possible role of this interaction in the expression of seed dormancy
Abstract
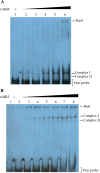
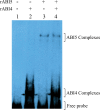
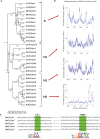
The precise adjustment of the timing of dormancy release according to final grain usage is still a challenge for many cereal crops. Grain sorghum [Sorghum bicolor (L.) Moench] shows wide intraspecific variability in dormancy level and susceptibility to pre-harvest sprouting (PHS). Both embryo sensitivity to abscisic acid (ABA) and gibberellin (GA) metabolism play an important role in the expression of dormancy of the developing sorghum grain. In previous works, it was shown that, simultaneously with a greater embryo sensitivity to ABA and higher expression of SbABA-INSENSITIVE 4 (SbABI4) and SbABA-INSENSITIVE 5 (SbABI5), dormant grains accumulate less active GA4 due to a more active GA catabolism. In this work, it is demonstrated that the ABA signalling components SbABI4 and SbABI5 interact in vitro with a fragment of the SbGA 2-OXIDASE 3 (SbGA2ox3) promoter containing an ABA-responsive complex (ABRC). Both transcription factors were able to bind the promoter, although not simultaneously, suggesting that they might compete for the same cis-acting regulatory sequences. A biological role for these interactions in the expression of dormancy of sorghum grains is proposed: either SbABI4 and/or SbABI5 activate transcription of the SbGA2ox3 gene in vivo and promote SbGA2ox3 protein accumulation; this would result in active degradation of GA4, thus preventing germination of dormant grains. A comparative analysis of the 5'-regulatory region of GA2oxs from both monocots and dicots is also presented; conservation of the ABRC in closely related GA2oxs from Brachypodium distachyon and rice suggest that these species might share the same regulatory mechanism as proposed for grain sorghum.
Keywords: ABI4; ABI5; GA 2-oxidase; Sorghum bicolor.; abscisic acid; germination; gibberellins; seed dormancy.
Figures


References
-
- Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, 28–36 - PubMed
-
- Barrero JM, Jacobsen JV, Talbot MJ, White RG, Swain SM, Garvin DF, Gubler F. 2012. Grain dormancy and light quality effects on germination in the model grass Brachypodium distachyon. New Phytologist 193, 376–386 - PubMed
-
- Baumlein H, Nagyt I, Villarroel R, Inzé D, Wobus U. 1992. Cis-analysis of a seed protein gene promoter: the conservative RY repeat CATGCATG within the legumin box is essential for tissue-specific expression of a legumin gene. The Plant Journal 2, 233–239 - PubMed
-
- Benech-Arnold RL, Enciso S, Sánchez RA, Carrari F, Perez-Flores L, Iusem N, Steinbach HS, Lijavetzky D, Bottini R. 2000. Involvement of ABA and GAs in the regulation of dormancy in developing sorghum seeds. In: Black M, Bradford KJ, Vázquez Ramos J, eds. Seed biology: advances and applications. Wallingford, UK, CAB International, 101–111
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

