Transcript processing and export kinetics are rate-limiting steps in expressing vertebrate segmentation clock genes
- PMID: 24151332
- PMCID: PMC3831944
- DOI: 10.1073/pnas.1308811110
Transcript processing and export kinetics are rate-limiting steps in expressing vertebrate segmentation clock genes
Abstract
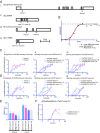
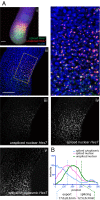
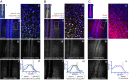
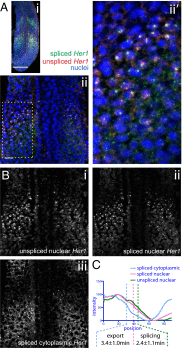
Sequential production of body segments in vertebrate embryos is regulated by a molecular oscillator (the segmentation clock) that drives cyclic transcription of genes involved in positioning intersegmental boundaries. Mathematical modeling indicates that the period of the clock depends on the total delay kinetics of a negative feedback circuit, including those associated with the synthesis of transcripts encoding clock components [Lewis J (2003) Curr Biol 13(16):1398-1408]. Here, we measure expression delays for three transcripts [Lunatic fringe, Hes7/her1, and Notch-regulated-ankyrin-repeat-protein (Nrarp)], that cycle during segmentation in the zebrafish, chick, and mouse, and provide in vivo measurements of endogenous splicing and export kinetics. We show that mRNA splicing and export are much slower than transcript elongation, with the longest delay (about 16 min in the mouse) being due to mRNA export. We conclude that the kinetics of mRNA and protein production and destruction can account for much of the clock period, and provide strong support for delayed autorepression as the underlying mechanism of the segmentation clock.
Keywords: RNA export; RNA splicing; mRNA processing; somites; transcriptional delays.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Cooke J, Zeeman EC. A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J Theor Biol. 1976;58(2):455–476. - PubMed
-
- Oates AC, Morelli LG, Ares S. Patterning embryos with oscillations: Structure, function and dynamics of the vertebrate segmentation clock. Development. 2012;139(4):625–639. - PubMed
-
- Palmeirim I, Henrique D, Ish-Horowicz D, Pourquié O. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell. 1997;91(5):639–648. - PubMed
-
- Jiang YJ, et al. Notch signalling and the synchronization of the somite segmentation clock. Nature. 2000;408(6811):475–479. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

