Structural basis for substrate specificity and mechanism of N-acetyl-D-neuraminic acid lyase from Pasteurella multocida
- PMID: 24152047
- PMCID: PMC3880309
- DOI: 10.1021/bi4011754
Structural basis for substrate specificity and mechanism of N-acetyl-D-neuraminic acid lyase from Pasteurella multocida
Abstract
N-Acetylneuraminate lyases (NALs) or sialic acid aldolases catalyze the reversible aldol cleavage of N-acetylneuraminic acid (Neu5Ac, the most common form of sialic acid) to form pyruvate and N-acetyl-d-mannosamine. Although equilibrium favors sialic acid cleavage, these enzymes can be used for high-yield chemoenzymatic synthesis of structurally diverse sialic acids in the presence of excess pyruvate. Engineering these enzymes to synthesize structurally modified natural sialic acids and their non-natural derivatives holds promise in creating novel therapeutic agents. Atomic-resolution structures of these enzymes will greatly assist in guiding mutagenic and modeling studies to engineer enzymes with altered substrate specificity. We report here the crystal structures of wild-type Pasteurella multocida N-acetylneuraminate lyase and its K164A mutant. Like other bacterial lyases, it assembles into a homotetramer with each monomer folding into a classic (β/α)₈ TIM barrel. Two wild-type structures were determined, in the absence of substrates, and trapped in a Schiff base intermediate between Lys164 and pyruvate, respectively. Three structures of the K164A variant were determined: one in the absence of substrates and two binary complexes with N-acetylneuraminic acid (Neu5Ac) and N-glycolylneuraminic acid (Neu5Gc). Both sialic acids bind to the active site in the open-chain ketone form of the monosaccharide. The structures reveal that every hydroxyl group of the linear sugars makes hydrogen bond interactions with the enzyme, and the residues that determine specificity were identified. Additionally, the structures provide some clues for explaining the natural discrimination of sialic acid substrates between the P. multocida and Escherichia coli NALs.
Figures




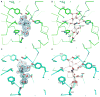
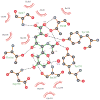
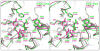
Similar articles
-
Crystal structures and kinetics of N-acetylneuraminate lyase from Fusobacterium nucleatum.Acta Crystallogr F Struct Biol Commun. 2018 Nov 1;74(Pt 11):725-732. doi: 10.1107/S2053230X18012992. Epub 2018 Oct 17. Acta Crystallogr F Struct Biol Commun. 2018. PMID: 30387778 Free PMC article.
-
Many locks to one key: N-acetylneuraminic acid binding to proteins.IUCrJ. 2024 Sep 1;11(Pt 5):664-674. doi: 10.1107/S2052252524005360. IUCrJ. 2024. PMID: 38965900 Free PMC article. Review.
-
Pasteurella multocida sialic acid aldolase: a promising biocatalyst.Appl Microbiol Biotechnol. 2008 Jul;79(6):963-70. doi: 10.1007/s00253-008-1506-2. Epub 2008 Jun 3. Appl Microbiol Biotechnol. 2008. PMID: 18521592 Free PMC article.
-
Molecular characterization of a novel N-acetylneuraminate lyase from Lactobacillus plantarum WCFS1.Appl Environ Microbiol. 2011 Apr;77(7):2471-8. doi: 10.1128/AEM.02927-10. Epub 2011 Feb 11. Appl Environ Microbiol. 2011. PMID: 21317263 Free PMC article.
-
The terminal enzymes of sialic acid metabolism: acylneuraminate pyruvate-lyases.Biosci Rep. 1999 Oct;19(5):373-83. doi: 10.1023/a:1020256004616. Biosci Rep. 1999. PMID: 10763805 Review.
Cited by
-
Features and structure of a cold active N-acetylneuraminate lyase.PLoS One. 2019 Jun 11;14(6):e0217713. doi: 10.1371/journal.pone.0217713. eCollection 2019. PLoS One. 2019. PMID: 31185017 Free PMC article.
-
First functional and mutational analysis of group 3 N-acetylneuraminate lyases from Lactobacillus antri and Lactobacillus sakei 23K.PLoS One. 2014 May 9;9(5):e96976. doi: 10.1371/journal.pone.0096976. eCollection 2014. PLoS One. 2014. PMID: 24817128 Free PMC article.
-
Crystal structures and kinetics of N-acetylneuraminate lyase from Fusobacterium nucleatum.Acta Crystallogr F Struct Biol Commun. 2018 Nov 1;74(Pt 11):725-732. doi: 10.1107/S2053230X18012992. Epub 2018 Oct 17. Acta Crystallogr F Struct Biol Commun. 2018. PMID: 30387778 Free PMC article.
-
Structure and function of bacterial transcription regulators of the SorC family.Transcription. 2024 Jun-Oct;15(3-5):139-160. doi: 10.1080/21541264.2024.2387895. Epub 2024 Sep 3. Transcription. 2024. PMID: 39223991 Free PMC article. Review.
-
Many locks to one key: N-acetylneuraminic acid binding to proteins.IUCrJ. 2024 Sep 1;11(Pt 5):664-674. doi: 10.1107/S2052252524005360. IUCrJ. 2024. PMID: 38965900 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

