Mapping genetic modifiers of survival in a mouse model of Dravet syndrome
- PMID: 24152123
- PMCID: PMC3930200
- DOI: 10.1111/gbb.12099
Mapping genetic modifiers of survival in a mouse model of Dravet syndrome
Abstract
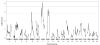
Epilepsy is a common neurological disorder affecting approximately 1% of the population. Mutations in voltage-gated sodium channels are responsible for several monogenic epilepsy syndromes. More than 800 mutations in the voltage-gated sodium channel SCN1A have been reported in patients with generalized epilepsy with febrile seizures plus and Dravet syndrome. Heterozygous loss-of-function mutations in SCN1A result in Dravet syndrome, a severe infant-onset epileptic encephalopathy characterized by intractable seizures, developmental delays and increased mortality. A common feature of monogenic epilepsies is variable expressivity among individuals with the same mutation, suggesting that genetic modifiers may influence clinical severity. Mice with heterozygous deletion of Scn1a (Scn1a(+/-) ) model a number of Dravet syndrome features, including spontaneous seizures and premature lethality. Phenotype severity in Scn1a(+/-) mice is strongly dependent on strain background. On the 129S6/SvEvTac strain Scn1a(+/-) mice exhibit no overt phenotype, whereas on the (C57BL/6J × 129S6/SvEvTac)F1 strain Scn1a(+/-) mice exhibit spontaneous seizures and early lethality. To systematically identify loci that influence premature lethality in Scn1a(+/-) mice, we performed genome scans on reciprocal backcrosses. Quantitative trait locus mapping revealed modifier loci on mouse chromosomes 5, 7, 8 and 11. RNA-seq analysis of strain-dependent gene expression, regulation and coding sequence variation provided a list of potential functional candidate genes at each locus. Identification of modifier genes that influence survival in Scn1a(+/-) mice will improve our understanding of the pathophysiology of Dravet syndrome and may suggest novel therapeutic strategies for improved treatment of human patients.
Keywords: Dravet syndrome; RNA-seq; epilepsy; epileptic encephalopathy; mouse model; seizures; severe myoclonic epilepsy of infancy; transcriptomics; voltage-gated ion channels; voltage-gated sodium channels.
© 2013 John Wiley & Sons Ltd and International Behavioural and Neural Genetics Society.
Figures
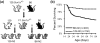


References
-
- Bergren SK, Chen S, Galecki A, Kearney JA. Genetic modifiers affecting severity of epilepsy caused by mutation of sodium channel Scn2a. Mamm Genome. 2005;16:683–690. - PubMed
-
- Bian F, Li Z, Offord J, Davis MD, McCormick J, Taylor CP, Walker LC. Calcium channel alpha2-delta type 1 subunit is the major binding protein for pregabalin in neocortex, hippocampus, amygdala, and spinal cord: an ex vivo autoradiographic study in alpha2-delta type 1 genetically modified mice. Brain Res. 2006;1075:68–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

