Further characterization of the target of a potential aptamer biomarker for pancreatic cancer: cyclophilin B and its posttranslational modifications
- PMID: 24152208
- PMCID: PMC3868381
- DOI: 10.1089/nat.2013.0439
Further characterization of the target of a potential aptamer biomarker for pancreatic cancer: cyclophilin B and its posttranslational modifications
Abstract
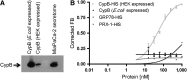
Posttranslational modifications on proteins can serve as useful biomarkers for disease. However, their discovery and detection in biological fluids is challenging. Aptamers are oligonucleotide ligands that demonstrate high affinity toward their target proteins and can discriminate closely related proteins with superb specificity. Previously, we generated a cyclophilin B aptamer (M9-5) that could discriminate sera from pancreatic cancer patients and healthy volunteers with high specificity and sensitivity. In our present work we further characterize the aptamer and the target protein, cyclophilin B, and demonstrate that the aptamer could discriminate between cyclophilin B expressed in human cells versus bacteria. Using mass-spectrometric analysis, we discovered post-translational modifications on cyclophilin B that might be responsible for the M9-5 selectivity. The ability to distinguish between forms of the same protein with differing post-translational modifications is an important advantage of aptamers as tools for identification and detection of biomarkers.
Figures

Similar articles
-
Comparing human pancreatic cell secretomes by in vitro aptamer selection identifies cyclophilin B as a candidate pancreatic cancer biomarker.J Clin Invest. 2012 May;122(5):1734-41. doi: 10.1172/JCI62385. Epub 2012 Apr 9. J Clin Invest. 2012. PMID: 22484812 Free PMC article.
-
The DNA aptamer binds stemness-enriched cancer cells in pancreatic cancer.J Mol Recognit. 2017 Apr;30(4). doi: 10.1002/jmr.2591. Epub 2016 Nov 28. J Mol Recognit. 2017. PMID: 27891685
-
Human Cyclophilin B Nuclease Activity Revealed via Nucleic Acid-Based Electrochemical Sensors.Angew Chem Int Ed Engl. 2022 Nov 7;61(45):e202211292. doi: 10.1002/anie.202211292. Epub 2022 Oct 6. Angew Chem Int Ed Engl. 2022. PMID: 35999181 Free PMC article.
-
Cancer protein biomarker discovery based on nucleic acid aptamers.Int J Biol Macromol. 2019 Jul 1;132:190-202. doi: 10.1016/j.ijbiomac.2019.03.165. Epub 2019 Mar 26. Int J Biol Macromol. 2019. PMID: 30926499 Review.
-
Post-translational modifications in tumor biomarkers: the next challenge for aptamers?Anal Bioanal Chem. 2018 Mar;410(8):2059-2065. doi: 10.1007/s00216-018-0861-9. Epub 2018 Jan 20. Anal Bioanal Chem. 2018. PMID: 29353432 Review.
Cited by
-
Hybrid-Type SELEX for the Selection of Artificial Nucleic Acid Aptamers Exhibiting Cell Internalization Activity.Pharmaceutics. 2021 Jun 15;13(6):888. doi: 10.3390/pharmaceutics13060888. Pharmaceutics. 2021. PMID: 34204006 Free PMC article.
-
In vitro selections of mammaglobin A and mammaglobin B aptamers for the recognition of circulating breast tumor cells.Sci Rep. 2017 Nov 3;7(1):14487. doi: 10.1038/s41598-017-13751-z. Sci Rep. 2017. PMID: 29101327 Free PMC article.
-
Development and classification of RNA aptamers for therapeutic purposes: an updated review with emphasis on cancer.Mol Cell Biochem. 2023 Jul;478(7):1573-1598. doi: 10.1007/s11010-022-04614-x. Epub 2022 Nov 24. Mol Cell Biochem. 2023. PMID: 36434145 Review.
-
Effects of codon optimization on coagulation factor IX translation and structure: Implications for protein and gene therapies.Sci Rep. 2019 Oct 29;9(1):15449. doi: 10.1038/s41598-019-51984-2. Sci Rep. 2019. PMID: 31664102 Free PMC article.
-
Trends in the Design and Development of Specific Aptamers Against Peptides and Proteins.Protein J. 2016 Apr;35(2):81-99. doi: 10.1007/s10930-016-9653-2. Protein J. 2016. PMID: 26984473 Review.
References
-
- ARIF M., SENAPATI P., SHANDILYA J., and KUNDU T.K. (2010). Protein lysine acetylation in cellular function and its role in cancer manifestation. Biochim. Biophys. Acta 1799,702–716 - PubMed
-
- BASKA K.M., MANANDHAR G., FENG D., AGCA Y., TENGOWSKI M.W., SUTOVSKY M., YI Y.J., and SUTOVSKY P. (2008). Mechanism of extracellular ubiquitination in the mammalian epididymis. J. Cell Physiol. 215,684–696 - PubMed
-
- BLOW N. (2007). Antibodies: the generation game. Nature 447,741–744 - PubMed
-
- CONRAD R., KERANEN L.M., ELLINGTON A.D., and NEWTON A.C. (1994). Isozyme-specific inhibition of protein kinase C by RNA aptamers. J. Biol. Chem. 269,32051–32054 - PubMed
-
- ELLINGTON A.D., and SZOSTAK J.W. (1990). In vitro selection of RNA molecules that bind specific ligands. Nature 346,818–822 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

