Mass spectrometry-based quantitative proteomics for dissecting multiplexed redox cysteine modifications in nitric oxide-protected cardiomyocyte under hypoxia
- PMID: 24152285
- PMCID: PMC3936484
- DOI: 10.1089/ars.2013.5326
Mass spectrometry-based quantitative proteomics for dissecting multiplexed redox cysteine modifications in nitric oxide-protected cardiomyocyte under hypoxia
Abstract
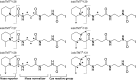
Aims: Distinctive states of redox-dependent cysteine (Cys) modifications are known to regulate signaling homeostasis under various pathophysiological conditions, including myocardial injury or protection in response to ischemic stress. Recent evidence further implicates a dynamic interplay among these modified forms following changes in cellular redox environment. However, a precise delineation of multiplexed Cys modifications in a cellular context remains technically challenging. To this end, we have now developed a mass spectrometry (MS)-based quantitative approach using a set of novel iodoacetyl-based Cys-reactive isobaric tags (irreversible isobaric iodoacetyl Cys-reactive tandem mass tag [iodoTMT]) endowed with unique irreversible Cys-reactivities.
Results: We have established a sequential iodoTMT-switch procedure coupled with efficient immunoenrichment and advanced shotgun liquid chromatography-MS/MS analysis. This workflow allows us to differentially quantify the multiple redox-modified forms of a Cys site in the original cellular context. In one single analysis, we have identified over 260 Cys sites showing quantitative differences in multiplexed redox modifications from the total lysates of H9c2 cardiomyocytes experiencing hypoxia in the absence and presence of S-nitrosoglutathione (GSNO), indicative of a distinct pattern of individual susceptibility to S-nitrosylation or S-glutathionylation. Among those most significantly affected are proteins functionally implicated in hypoxic damage from which we showed that GSNO would protect.
Innovation: We demonstrate for the first time how quantitative analysis of various Cys-redox modifications occurring in biological samples can be performed precisely and simultaneously at proteomic levels.
Conclusion: We have not only developed a new approach to map global Cys-redoxomic regulation in vivo, but also provided new evidences implicating Cys-redox modifications of key molecules in NO-mediated ischemic cardioprotection.
Figures






References
-
- Aracena P, Tang W, Hamilton SL, and Hidalgo C. Effects of S-glutathionylation and S-nitrosylation on calmodulin binding to triads and FKBP12 binding to type 1 calcium release channels. Antioxid Redox Signal 7: 870–881, 2005 - PubMed
-
- Aracena-Parks P, Goonasekera SA, Gilman CP, Dirksen RT, Hidalgo C, and Hamilton SL. Identification of cysteines involved in S-nitrosylation, S-glutathionylation, and oxidation to disulfides in ryanodine receptor type 1. J Biol Chem 281: 40354–40368, 2006 - PubMed
-
- Becker LB. New concepts in reactive oxygen species and cardiovascular reperfusion physiology. Cardiovasc Res 61: 461–470, 2004 - PubMed
-
- Brennan JP, Miller JI, Fuller W, Wait R, Begum S, Dunn MJ, and Eaton P. The utility of N,N-biotinyl glutathione disulfide in the study of protein S-glutathiolation. Mol Cell Proteomics 5: 215–225, 2006 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

