A novel taxonomic marker that discriminates between morphologically complex actinomycetes
- PMID: 24153003
- PMCID: PMC3814722
- DOI: 10.1098/rsob.130073
A novel taxonomic marker that discriminates between morphologically complex actinomycetes
Abstract
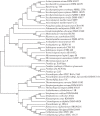
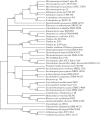
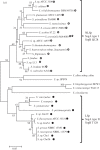
In the era when large whole genome bacterial datasets are generated routinely, rapid and accurate molecular systematics is becoming increasingly important. However, 16S ribosomal RNA sequencing does not always offer sufficient resolution to discriminate between closely related genera. The SsgA-like proteins are developmental regulatory proteins in sporulating actinomycetes, whereby SsgB actively recruits FtsZ during sporulation-specific cell division. Here, we present a novel method to classify actinomycetes, based on the extraordinary way the SsgA and SsgB proteins are conserved. The almost complete conservation of the SsgB amino acid (aa) sequence between members of the same genus and its high divergence between even closely related genera provides high-quality data for the classification of morphologically complex actinomycetes. Our analysis validates Kitasatospora as a sister genus to Streptomyces in the family Streptomycetaceae and suggests that Micromonospora, Salinispora and Verrucosispora may represent different clades of the same genus. It is also apparent that the aa sequence of SsgA is an accurate determinant for the ability of streptomycetes to produce submerged spores, dividing the phylogenetic tree of streptomycetes into liquid-culture sporulation and no liquid-culture sporulation branches. A new phylogenetic tree of industrially relevant actinomycetes is presented and compared with that based on 16S rRNA sequences.
Keywords: Streptomyces; cell division; genome sequencing; systematics.
Figures



Similar articles
-
The SsgA-like proteins in actinomycetes: small proteins up to a big task.Antonie Van Leeuwenhoek. 2008 Jun;94(1):85-97. doi: 10.1007/s10482-008-9225-3. Epub 2008 Feb 14. Antonie Van Leeuwenhoek. 2008. PMID: 18273689 Free PMC article. Review.
-
Structural and functional characterizations of SsgB, a conserved activator of developmental cell division in morphologically complex actinomycetes.J Biol Chem. 2009 Sep 11;284(37):25268-79. doi: 10.1074/jbc.M109.018564. Epub 2009 Jun 30. J Biol Chem. 2009. PMID: 19567872 Free PMC article.
-
Analysis of novel kitasatosporae reveals significant evolutionary changes in conserved developmental genes between Kitasatospora and Streptomyces.Antonie Van Leeuwenhoek. 2014 Aug;106(2):365-80. doi: 10.1007/s10482-014-0209-1. Epub 2014 Jun 24. Antonie Van Leeuwenhoek. 2014. PMID: 24958203
-
Characterization of the sporulation control protein SsgA by use of an efficient method to create and screen random mutant libraries in streptomycetes.Appl Environ Microbiol. 2007 Apr;73(7):2085-92. doi: 10.1128/AEM.02755-06. Epub 2007 Feb 9. Appl Environ Microbiol. 2007. PMID: 17293502 Free PMC article.
-
Genus Kitasatospora, taxonomic features and diversity of secondary metabolites.J Antibiot (Tokyo). 2017 May;70(5):506-513. doi: 10.1038/ja.2017.8. Epub 2017 Feb 15. J Antibiot (Tokyo). 2017. PMID: 28196972 Review.
Cited by
-
Sporulation-specific cell division defects in ylmE mutants of Streptomyces coelicolor are rescued by additional deletion of ylmD.Sci Rep. 2018 May 9;8(1):7328. doi: 10.1038/s41598-018-25782-1. Sci Rep. 2018. PMID: 29743540 Free PMC article.
-
Marine Actinomycetes, New Sources of Biotechnological Products.Mar Drugs. 2021 Jun 25;19(7):365. doi: 10.3390/md19070365. Mar Drugs. 2021. PMID: 34201951 Free PMC article. Review.
-
Adaptation to Endophytic Lifestyle Through Genome Reduction by Kitasatospora sp. SUK42.Front Bioeng Biotechnol. 2021 Oct 12;9:740722. doi: 10.3389/fbioe.2021.740722. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 34712653 Free PMC article.
-
Isolation of β-1,3-Glucanase-Producing Microorganisms from Poria cocos Cultivation Soil via Molecular Biology.Molecules. 2018 Jun 27;23(7):1555. doi: 10.3390/molecules23071555. Molecules. 2018. PMID: 29954113 Free PMC article.
-
Diversity and functions of volatile organic compounds produced by Streptomyces from a disease-suppressive soil.Front Microbiol. 2015 Oct 9;6:1081. doi: 10.3389/fmicb.2015.01081. eCollection 2015. Front Microbiol. 2015. PMID: 26500626 Free PMC article.
References
-
- Shendure J, Lieberman Aiden E. 2012. The expanding scope of DNA sequencing. Nat. Biotechnol. 30, 1084–1094 (doi:10.1038/nbt.2421) - DOI - PMC - PubMed
-
- Goodfellow M. 2012. Phylum XXVI. Actinobacteria phyl. nov. In Bergey's manual of systematic bacteriology, 2nd edn (eds Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W, Whitman WB.), pp. 1–2083 New York, NY: Springer
-
- Bérdy J. 2012. Thoughts and facts about antibiotics: where we are now and where we are heading. J. Antibiot. (Tokyo) 65, 385–395 (doi:10.1038/ja.2012.27) - DOI - PubMed
-
- Hopwood DA. 2007. Streptomyces in nature and medicine: the antibiotic makers. New York, NY: Oxford University Press
-
- Bentley SD, et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147 (doi:10.1038/417141a) - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

