A novel taxonomic marker that discriminates between morphologically complex actinomycetes
- PMID: 24153003
- PMCID: PMC3814722
- DOI: 10.1098/rsob.130073
A novel taxonomic marker that discriminates between morphologically complex actinomycetes
Abstract
In the era when large whole genome bacterial datasets are generated routinely, rapid and accurate molecular systematics is becoming increasingly important. However, 16S ribosomal RNA sequencing does not always offer sufficient resolution to discriminate between closely related genera. The SsgA-like proteins are developmental regulatory proteins in sporulating actinomycetes, whereby SsgB actively recruits FtsZ during sporulation-specific cell division. Here, we present a novel method to classify actinomycetes, based on the extraordinary way the SsgA and SsgB proteins are conserved. The almost complete conservation of the SsgB amino acid (aa) sequence between members of the same genus and its high divergence between even closely related genera provides high-quality data for the classification of morphologically complex actinomycetes. Our analysis validates Kitasatospora as a sister genus to Streptomyces in the family Streptomycetaceae and suggests that Micromonospora, Salinispora and Verrucosispora may represent different clades of the same genus. It is also apparent that the aa sequence of SsgA is an accurate determinant for the ability of streptomycetes to produce submerged spores, dividing the phylogenetic tree of streptomycetes into liquid-culture sporulation and no liquid-culture sporulation branches. A new phylogenetic tree of industrially relevant actinomycetes is presented and compared with that based on 16S rRNA sequences.
Keywords: Streptomyces; cell division; genome sequencing; systematics.
Figures

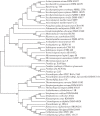
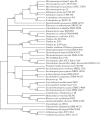

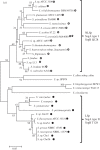

References
-
- Shendure J, Lieberman Aiden E. 2012. The expanding scope of DNA sequencing. Nat. Biotechnol. 30, 1084–1094 (doi:10.1038/nbt.2421) - DOI - PMC - PubMed
-
- Goodfellow M. 2012. Phylum XXVI. Actinobacteria phyl. nov. In Bergey's manual of systematic bacteriology, 2nd edn (eds Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W, Whitman WB.), pp. 1–2083 New York, NY: Springer
-
- Bérdy J. 2012. Thoughts and facts about antibiotics: where we are now and where we are heading. J. Antibiot. (Tokyo) 65, 385–395 (doi:10.1038/ja.2012.27) - DOI - PubMed
-
- Hopwood DA. 2007. Streptomyces in nature and medicine: the antibiotic makers. New York, NY: Oxford University Press
-
- Bentley SD, et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147 (doi:10.1038/417141a) - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

