Comparison of sputum and nasopharyngeal aspirate samples and of the PCR gene targets lytA and Spn9802 for quantitative PCR for rapid detection of pneumococcal pneumonia
- PMID: 24153121
- PMCID: PMC3911438
- DOI: 10.1128/JCM.01742-13
Comparison of sputum and nasopharyngeal aspirate samples and of the PCR gene targets lytA and Spn9802 for quantitative PCR for rapid detection of pneumococcal pneumonia
Abstract
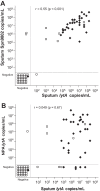
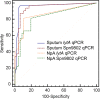
We aimed to compare sputum and nasopharyngeal aspirate (NpA) samples and the PCR gene targets lytA and Spn9802 in quantitative PCR (qPCR) assays for rapid detection of pneumococcal etiology in community-acquired pneumonia (CAP). Seventy-eight adult patients hospitalized for radiologically confirmed CAP had both good-quality sputum and NpA specimens collected at admission. These samples were subjected to lytA qPCR and Spn9802 qPCR assays with analytical times of <3 h. Thirty-two patients had CAP with a pneumococcal etiology, according to conventional diagnostic criteria. The following qPCR positivity rates were noted in CAP cases with and without pneumococcal etiology: 96% and 15% (sputum lytA assay), 96% and 17% (sputum Spn9802 assay), 81% and 11% (NpA lytA assay), and 81% and 20% (NpA Spn9802 assay), respectively. The mean lytA and Spn9802 DNA levels were significantly higher in qPCR-positive sputum samples from cases with pneumococcal etiology than in qPCR-positive sputum samples from CAP cases without pneumococcal etiology or qPCR-positive NpA samples from cases with pneumococcal etiology (P < 0.02 for all comparisons). For detection of pneumococcal etiology, receiver operating characteristic curve analysis showed that sputum specimens were superior to NpA specimens as the sample type (P < 0.02 for both gene targets) and lytA tended to be superior to Spn9802 as the gene target. The best-performing test, the sputum lytA qPCR assay, showed high sensitivity (94%) and specificity (96%) with a cutoff value of 10(5) DNA copies/ml. In CAP patients with good sputum production, this test has great potential to be used for the rapid detection of pneumococcal etiology and to target penicillin therapy.
Figures

References
-
- Spindler C, Strålin K, Eriksson L, Hjerdt-Goscinski G, Holmberg H, Lidman C, Nilsson A, Örtqvist Å, Hedlund J. 2012. Swedish guidelines on the management of community-acquired pneumonia in immunocompetent adults: Swedish Society of Infectious Diseases 2012. Scand. J. Infect. Dis. 44:885–902. 10.3109/00365548.2012.700120 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

