Differential L1 regulation in pluripotent stem cells of humans and apes
- PMID: 24153179
- PMCID: PMC4064720
- DOI: 10.1038/nature12686
Differential L1 regulation in pluripotent stem cells of humans and apes
Abstract
Identifying cellular and molecular differences between human and non-human primates (NHPs) is essential to the basic understanding of the evolution and diversity of our own species. Until now, preserved tissues have been the main source for most comparative studies between humans, chimpanzees (Pan troglodytes) and bonobos (Pan paniscus). However, these tissue samples do not fairly represent the distinctive traits of live cell behaviour and are not amenable to genetic manipulation. We propose that induced pluripotent stem (iPS) cells could be a unique biological resource to determine relevant phenotypical differences between human and NHPs, and that those differences could have potential adaptation and speciation value. Here we describe the generation and initial characterization of iPS cells from chimpanzees and bonobos as new tools to explore factors that may have contributed to great ape evolution. Comparative gene expression analysis of human and NHP iPS cells revealed differences in the regulation of long interspersed element-1 (L1, also known as LINE-1) transposons. A force of change in mammalian evolution, L1 elements are retrotransposons that have remained active during primate evolution. Decreased levels of L1-restricting factors APOBEC3B (also known as A3B) and PIWIL2 (ref. 7) in NHP iPS cells correlated with increased L1 mobility and endogenous L1 messenger RNA levels. Moreover, results from the manipulation of A3B and PIWIL2 levels in iPS cells supported a causal inverse relationship between levels of these proteins and L1 retrotransposition. Finally, we found increased copy numbers of species-specific L1 elements in the genome of chimpanzees compared to humans, supporting the idea that increased L1 mobility in NHPs is not limited to iPS cells in culture and may have also occurred in the germ line or embryonic cells developmentally upstream to germline specification during primate evolution. We propose that differences in L1 mobility may have differentially shaped the genomes of humans and NHPs and could have continuing adaptive significance.
Figures
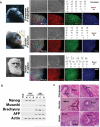
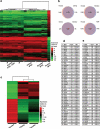

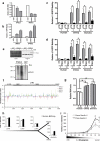


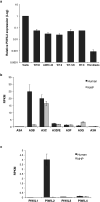



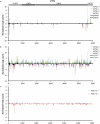
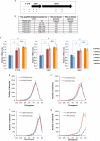
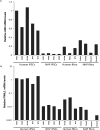
Comment in
-
Human evolution: reprogrammed cells dissect ape retrotransposition.Nat Rev Genet. 2013 Dec;14(12):824. doi: 10.1038/nrg3621. Epub 2013 Nov 12. Nat Rev Genet. 2013. PMID: 24217322 No abstract available.
References
-
- Chimpanzee S, Analysis C. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. - PubMed
-
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. - PubMed
-
- Kazazian HH., Jr. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi:10.1126/science.1089670. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- R01 MH095741/MH/NIMH NIH HHS/United States
- AI074967/AI/NIAID NIH HHS/United States
- R01 GM084317/GM/NIGMS NIH HHS/United States
- NS075449/NS/NINDS NIH HHS/United States
- R01 MH094753/MH/NIMH NIH HHS/United States
- R01 HG004659/HG/NHGRI NIH HHS/United States
- R01 MH088485/MH/NIMH NIH HHS/United States
- P30 CA014195/CA/NCI NIH HHS/United States
- HG004659/HG/NHGRI NIH HHS/United States
- MH094753/MH/NIMH NIH HHS/United States
- MH08848/MH/NIMH NIH HHS/United States
- GM084317/GM/NIGMS NIH HHS/United States
- R01 NS075449/NS/NINDS NIH HHS/United States
- R01 AI074967/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

