IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication
- PMID: 24154727
- PMCID: PMC3845190
- DOI: 10.1073/pnas.1311669110
IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication
Erratum in
- Proc Natl Acad Sci U S A. 2013 Nov 26;110(48):19651. Jin, Tengchuan [corrected to Tengchuan, Jin]
Abstract
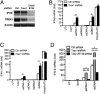
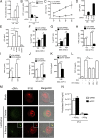
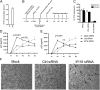
Replication of lentiviruses generates different DNA forms, including RNA:DNA hybrids, ssDNA, and dsDNA. Nucleic acids stimulate innate immune responses, and pattern recognition receptors detecting dsDNA have been identified. However, sensors for ssDNA have not been reported, and the ability of RNA:DNA hybrids to stimulate innate immune responses is controversial. Using ssDNAs derived from HIV-1 proviral DNA, we report that this DNA form potently induces the expression of IFNs in primary human macrophages. This response was stimulated by stem regions in the DNA structure and was dependent on IFN-inducible protein 16 (IFI16), which bound immunostimulatory DNA directly and activated the stimulator of IFN genes -TANK-binding kinase 1 - IFN regulatory factors 3/7 (STING-TBK1-IRF3/7) pathway. Importantly, IFI16 colocalized and associated with lentiviral DNA in the cytoplasm in macrophages, and IFI16 knockdown in this cell type augmented lentiviral transduction and also HIV-1 replication. Thus, IFI16 is a sensor for DNA forms produced during the lentiviral replication cycle and regulates HIV-1 replication in macrophages.
Keywords: DNA sensing; antiviral defense; innate immunity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Comment in
-
HIV provides ample PAMPs for innate immune sensing.Proc Natl Acad Sci U S A. 2013 Nov 26;110(48):19183-4. doi: 10.1073/pnas.1319118110. Epub 2013 Nov 13. Proc Natl Acad Sci U S A. 2013. PMID: 24225850 Free PMC article. No abstract available.
References
-
- Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–105. - PubMed
-
- Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448(7152):501–505. - PubMed
-
- Heil F, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303(5663):1526–1529. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

