Activation of influenza A viruses by host proteases from swine airway epithelium
- PMID: 24155384
- PMCID: PMC3911738
- DOI: 10.1128/JVI.01635-13
Activation of influenza A viruses by host proteases from swine airway epithelium
Abstract
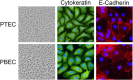
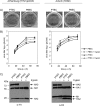
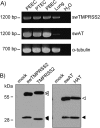
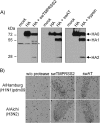
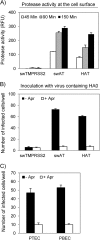
Pigs are important natural hosts of influenza A viruses, and due to their susceptibility to swine, avian, and human viruses, they may serve as intermediate hosts supporting adaptation and genetic reassortment. Cleavage of the influenza virus surface glycoprotein hemagglutinin (HA) by host cell proteases is essential for viral infectivity. Most influenza viruses, including human and swine viruses, are activated at a monobasic HA cleavage site, and we previously identified TMPRSS2 and HAT to be relevant proteases present in human airways. We investigated the proteolytic activation of influenza viruses in primary porcine tracheal and bronchial epithelial cells (PTEC and PBEC, respectively). Human H1N1 and H3N2 viruses replicated efficiently in PTECs and PBECs, and viruses containing cleaved HA were released from infected cells. Moreover, the cells supported the proteolytic activation of HA at the stage of entry. We found that swine proteases homologous to TMPRSS2 and HAT, designated swTMPRSS2 and swAT, respectively, were expressed in several parts of the porcine respiratory tract. Both proteases cloned from primary PBECs were shown to activate HA with a monobasic cleavage site upon coexpression and support multicycle replication of influenza viruses. swAT was predominantly localized at the plasma membrane, where it was present as an active protease that mediated activation of incoming virus. In contrast, swTMPRSS2 accumulated in the trans-Golgi network, suggesting that it cleaves HA in this compartment. In conclusion, our data show that HA activation in porcine airways may occur by similar proteases and at similar stages of the viral life cycle as in human airways.
Figures
Similar articles
-
Hemagglutinins of Avian Influenza Viruses Are Proteolytically Activated by TMPRSS2 in Human and Murine Airway Cells.J Virol. 2021 Sep 27;95(20):e0090621. doi: 10.1128/JVI.00906-21. Epub 2021 Jul 28. J Virol. 2021. PMID: 34319155 Free PMC article.
-
TMPRSS2 is a host factor that is essential for pneumotropism and pathogenicity of H7N9 influenza A virus in mice.J Virol. 2014 May;88(9):4744-51. doi: 10.1128/JVI.03799-13. Epub 2014 Feb 12. J Virol. 2014. PMID: 24522916 Free PMC article.
-
Transcriptome profiling and protease inhibition experiments identify proteases that activate H3N2 influenza A and influenza B viruses in murine airways.J Biol Chem. 2020 Aug 14;295(33):11388-11407. doi: 10.1074/jbc.RA120.012635. Epub 2020 Apr 17. J Biol Chem. 2020. PMID: 32303635 Free PMC article.
-
Activation of influenza viruses by proteases from host cells and bacteria in the human airway epithelium.Pathog Dis. 2013 Nov;69(2):87-100. doi: 10.1111/2049-632X.12053. Epub 2013 Jul 2. Pathog Dis. 2013. PMID: 23821437 Free PMC article. Review.
-
Novel insights into proteolytic cleavage of influenza virus hemagglutinin.Rev Med Virol. 2010 Sep;20(5):298-310. doi: 10.1002/rmv.657. Rev Med Virol. 2010. PMID: 20629046 Free PMC article. Review.
Cited by
-
The role of influenza-A virus and coronavirus viral glycoprotein cleavage in host adaptation.Curr Opin Virol. 2023 Feb;58:101303. doi: 10.1016/j.coviro.2023.101303. Epub 2023 Jan 18. Curr Opin Virol. 2023. PMID: 36753938 Free PMC article. Review.
-
The serine protease inhibitor camostat inhibits influenza virus replication and cytokine production in primary cultures of human tracheal epithelial cells.Pulm Pharmacol Ther. 2015 Aug;33:66-74. doi: 10.1016/j.pupt.2015.07.001. Epub 2015 Jul 10. Pulm Pharmacol Ther. 2015. PMID: 26166259 Free PMC article.
-
Identification of a broad-spectrum antiviral small molecule against severe acute respiratory syndrome coronavirus and Ebola, Hendra, and Nipah viruses by using a novel high-throughput screening assay.J Virol. 2014 Apr;88(8):4353-65. doi: 10.1128/JVI.03050-13. Epub 2014 Feb 5. J Virol. 2014. PMID: 24501399 Free PMC article.
-
Equine and Canine Influenza H3N8 Viruses Show Minimal Biological Differences Despite Phylogenetic Divergence.J Virol. 2015 Jul;89(13):6860-73. doi: 10.1128/JVI.00521-15. Epub 2015 Apr 22. J Virol. 2015. PMID: 25903329 Free PMC article.
-
An overview of influenza A virus genes, protein functions, and replication cycle highlighting important updates.Virus Genes. 2022 Aug;58(4):255-269. doi: 10.1007/s11262-022-01904-w. Epub 2022 Apr 26. Virus Genes. 2022. PMID: 35471490 Review.
References
-
- Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204. 10.1126/science.1222213 - DOI - PMC - PubMed
-
- Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. 2012. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. U. S. A. 109:4269–4274. 10.1073/pnas.1116200109 - DOI - PMC - PubMed
-
- Matrosovich M, Stech J, Klenk HD. 2009. Influenza receptors, polymerase and host range. Rev. Sci. Tech. 28:203–217 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

