Differential accessibility of a rotavirus VP6 epitope in trimers comprising type I, II, or III channels as revealed by binding of a human rotavirus VP6-specific antibody
- PMID: 24155406
- PMCID: PMC3911710
- DOI: 10.1128/JVI.01665-13
Differential accessibility of a rotavirus VP6 epitope in trimers comprising type I, II, or III channels as revealed by binding of a human rotavirus VP6-specific antibody
Abstract
Previous human antibody studies have shown that the human VH1-46 antibody variable gene segment encodes much of the naturally occurring human B cell response to rotavirus and is directed to virus protein 6 (VP6). It is currently unknown why some of the VH1-46-encoded human VP6 monoclonal antibodies inhibit viral transcription while others do not. In part, there are affinity differences between antibodies that likely affect inhibitory activity, but we also hypothesize that there are differing modes of binding to VP6 that affect the ability to block the transcriptional pore on double-layered particles. Here, we used a hybrid method approach for antibody epitope mapping, including single-particle cryo-electron microscopy (cryo-EM) and enhanced amide hydrogen-deuterium exchange mass spectrometry (DXMS) to determine the location and mode of binding of a VH1-46-encoded antibody, RV6-25. The structure of the RV6-25 antibody-double-layered particle (DLP) complex indicated a very complex binding pattern that revealed subtle differences in accessibility of the VP6 epitope depending on its position in the type I, II, or III channels. These subtle variations in the presentation or accessibility of the RV VP6 capsid layer led to position-specific differences in occupancy for binding of the RV6-25 antibody. The studies also showed that the location of binding of the noninhibitory antibody RV6-25 on the apical surface of RV VP6 head domain does not obstruct the transcription pore upon antibody binding, in contrast to binding of an inhibitory antibody, RV6-26, deeper in the transcriptional pore.
Figures

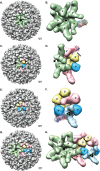

Similar articles
-
Human rotavirus VP6-specific antibodies mediate intracellular neutralization by binding to a quaternary structure in the transcriptional pore.PLoS One. 2013 May 9;8(5):e61101. doi: 10.1371/journal.pone.0061101. Print 2013. PLoS One. 2013. PMID: 23671563 Free PMC article.
-
Antibody inhibition of the transcriptase activity of the rotavirus DLP: a structural view.J Mol Biol. 2001 Mar 16;307(1):161-72. doi: 10.1006/jmbi.2000.4479. J Mol Biol. 2001. PMID: 11243811
-
Identification of rotavirus VP6 residues located at the interface with VP2 that are essential for capsid assembly and transcriptase activity.J Virol. 2002 Aug;76(15):7822-31. doi: 10.1128/jvi.76.15.7822-7831.2002. J Virol. 2002. PMID: 12097594 Free PMC article.
-
Rotavirus VP6 preparations as a non-replicating vaccine candidates.Vaccine. 2015 Jun 26;33(29):3281-7. doi: 10.1016/j.vaccine.2015.05.026. Epub 2015 May 26. Vaccine. 2015. PMID: 26021725 Review.
-
VP6: A candidate rotavirus vaccine.J Infect Dis. 2010 Sep 1;202 Suppl:S101-7. doi: 10.1086/653556. J Infect Dis. 2010. PMID: 20684688 Review.
Cited by
-
Intragenic recombination influences rotavirus diversity and evolution.Virus Evol. 2020 Jan 13;6(1):vez059. doi: 10.1093/ve/vez059. eCollection 2020 Jan. Virus Evol. 2020. PMID: 31949920 Free PMC article.
-
Characterization of a genetically heterogeneous porcine rotavirus C, and other viruses present in the fecal virome of a non-diarrheic Belgian piglet.Infect Genet Evol. 2016 Sep;43:135-45. doi: 10.1016/j.meegid.2016.05.018. Epub 2016 May 14. Infect Genet Evol. 2016. PMID: 27184192 Free PMC article.
-
High Resolution Mapping of Bactericidal Monoclonal Antibody Binding Epitopes on Staphylococcus aureus Antigen MntC.PLoS Pathog. 2016 Sep 30;12(9):e1005908. doi: 10.1371/journal.ppat.1005908. eCollection 2016 Sep. PLoS Pathog. 2016. PMID: 27689696 Free PMC article.
-
Spectroscopic analysis of the bacterially expressed head domain of rotavirus VP6.Biosci Rep. 2024 May 29;44(5):BSR20232178. doi: 10.1042/BSR20232178. Biosci Rep. 2024. PMID: 38592735 Free PMC article.
-
A Site of Vulnerability on the Influenza Virus Hemagglutinin Head Domain Trimer Interface.Cell. 2019 May 16;177(5):1136-1152.e18. doi: 10.1016/j.cell.2019.04.011. Cell. 2019. PMID: 31100268 Free PMC article.
References
-
- Estes MK. 2001. Rotavirus and their replication, p 1747–1785 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 4th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA
-
- Estes MK, Kapikian AZ. 2007. Rotaviruses, p 1918–1974 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

