DNA methylation profiling revealed promoter hypermethylation-induced silencing of p16, DDAH2 and DUSP1 in primary oral squamous cell carcinoma
- PMID: 24155659
- PMCID: PMC3805925
- DOI: 10.7150/ijms.6884
DNA methylation profiling revealed promoter hypermethylation-induced silencing of p16, DDAH2 and DUSP1 in primary oral squamous cell carcinoma
Abstract
Background: Hypermethylation in promoter regions of genes might lead to altered gene functions and result in malignant cellular transformation. Thus, biomarker identification for hypermethylated genes would be very useful for early diagnosis, prognosis, and therapeutic treatment of oral squamous cell carcinoma (OSCC). The objectives of this study were to screen and validate differentially hypermethylated genes in OSCC and correlate the hypermethylation-induced genes with demographic, clinocopathological characteristics and survival rate of OSCC.
Methods: DNA methylation profiling was utilized to screen the differentially hypermethylated genes in OSCC. Three selected differentially-hypermethylated genes of p16, DDAH2 and DUSP1 were further validated for methylation status and protein expression. The correlation between demographic, clinicopathological characteristics, and survival rate of OSCC patients with hypermethylation of p16, DDAH2 and DUSP1 genes were analysed in the study.
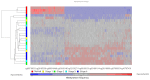
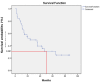
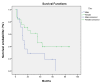
Results: Methylation profiling demonstrated 33 promoter hypermethylated genes in OSCC. The differentially-hypermethylated genes of p16, DDAH2 and DUSP1 revealed positivity of 78%, 80% and 88% in methylation-specific polymerase chain reaction and 24% and 22% of immunoreactivity in DDAH2 and DUSP1 genes, respectively. Promoter hypermethylation of p16 gene was found significantly associated with tumour site of buccal, gum, tongue and lip (P=0.001). In addition, DDAH2 methylation level was correlated significantly with patients' age (P=0.050). In this study, overall five-year survival rate was 38.1% for OSCC patients and was influenced by sex difference.
Conclusions: The study has identified 33 promoter hypermethylated genes that were significantly silenced in OSCC, which might be involved in an important mechanism in oral carcinogenesis. Our approaches revealed signature candidates of differentially hypermethylated genes of DDAH2 and DUSP1 which can be further developed as potential biomarkers for OSCC as diagnostic, prognostic and therapeutic targets in the future.
Keywords: DDAH2; DNA methylation profiling; DUSP1; Oral Squamous Cell Carcinoma; promoter hypermethylation; protein expression..
Conflict of interest statement
Competing interests: The authors have declared that no competing interest.
Figures






Similar articles
-
PTEN and p16 genes as epigenetic biomarkers in oral squamous cell carcinoma (OSCC): a study on south Indian population.Tumour Biol. 2016 Jun;37(6):7625-32. doi: 10.1007/s13277-015-4648-8. Epub 2015 Dec 19. Tumour Biol. 2016. PMID: 26687648
-
Screening of differential promoter hypermethylated genes in primary oral squamous cell carcinoma.Asian Pac J Cancer Prev. 2014;15(20):8957-61. doi: 10.7314/apjcp.2014.15.20.8957. Asian Pac J Cancer Prev. 2014. PMID: 25374236
-
Aberrantly hypermethylated tumor suppressor genes were identified in oral squamous cell carcinoma (OSCC).Clin Epigenetics. 2019 Aug 12;11(1):116. doi: 10.1186/s13148-019-0715-0. Clin Epigenetics. 2019. PMID: 31405379 Free PMC article.
-
p16INK4A and p14ARF gene promoter hypermethylation as prognostic biomarker in oral and oropharyngeal squamous cell carcinoma: a review.Dis Markers. 2014;2014:260549. doi: 10.1155/2014/260549. Epub 2014 Apr 7. Dis Markers. 2014. PMID: 24803719 Free PMC article. Review.
-
DNA hypermethylation of tumor suppressor genes among oral squamous cell carcinoma patients: a prominent diagnostic biomarker.Mol Biol Rep. 2024 Dec 7;52(1):44. doi: 10.1007/s11033-024-10144-0. Mol Biol Rep. 2024. PMID: 39644423 Review.
Cited by
-
DNA methylation and obesity traits: An epigenome-wide association study. The REGICOR study.Epigenetics. 2017;12(10):909-916. doi: 10.1080/15592294.2017.1363951. Epub 2017 Nov 27. Epigenetics. 2017. PMID: 29099282 Free PMC article.
-
Portrait of DNA methylated genes predictive of poor prognosis in head and neck cancer and the implication for targeted therapy.Sci Rep. 2021 May 11;11(1):10012. doi: 10.1038/s41598-021-89476-x. Sci Rep. 2021. PMID: 33976322 Free PMC article.
-
Distribution of PAX1 and ZNF582 Hypermethylation in the Oral Exfoliated Cells of Oral Squamous Cell Carcinoma.Clin Med Insights Oncol. 2025 May 16;19:11795549251335172. doi: 10.1177/11795549251335172. eCollection 2025. Clin Med Insights Oncol. 2025. PMID: 40386310 Free PMC article.
-
DUSP1 is involved in the progression of small cell carcinoma of the prostate.Saudi J Biol Sci. 2018 Jul;25(5):858-862. doi: 10.1016/j.sjbs.2017.09.015. Epub 2017 Oct 18. Saudi J Biol Sci. 2018. PMID: 30108432 Free PMC article.
-
An integrated methylation and gene expression microarray analysis reveals significant prognostic biomarkers in oral squamous cell carcinoma.Oncol Rep. 2018 Nov;40(5):2637-2647. doi: 10.3892/or.2018.6702. Epub 2018 Sep 12. Oncol Rep. 2018. PMID: 30226546 Free PMC article.
References
-
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA: a cancer journal for clinicians. 2005;55:74–108. - PubMed
-
- Jemal A, Bray F, Center MM. et al. Global cancer statistics. CA: a cancer journal for clinicians. 2011;61:69–90. - PubMed
-
- Kimman M, Norman R, Jan S. et al. The burden of cancer in member countries of the Association of Southeast Asian Nations (ASEAN) Asian Pac J Cancer Prev. 2012;13:411–420. - PubMed
-
- Kim TJ, Kim HY, Lee KW, Kim MS. Multimodality Assessment of Esophageal Cancer: Preoperative Staging and Monitoring of Response to Therapy. Radiographics. 2009;29:403–421. - PubMed
-
- Mulero-Navarro S, Esteller M. Epigenetic biomarkers for human cancer: The time is now. Critical Reviews in Oncology/Hematology. 2008;68:1–11. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

