Improved PCR performance using template DNA from formalin-fixed and paraffin-embedded tissues by overcoming PCR inhibition
- PMID: 24155973
- PMCID: PMC3796491
- DOI: 10.1371/journal.pone.0077771
Improved PCR performance using template DNA from formalin-fixed and paraffin-embedded tissues by overcoming PCR inhibition
Abstract
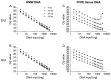
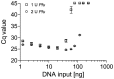
Formalin-fixed and paraffin-embedded (FFPE) tissues represent a valuable source for biomarker studies and clinical routine diagnostics. However, they suffer from degradation of nucleic acids due to the fixation process. Since genetic and epigenetic studies usually require PCR amplification, this degradation hampers its use significantly, impairing PCR robustness or necessitating short amplicons. In routine laboratory medicine a highly robust PCR performance is mandatory for the clinical utility of genetic and epigenetic biomarkers. Therefore, methods to improve PCR performance using DNA from FFPE tissue are highly desired and of wider interest. The effect of template DNA derived from FFPE tissues on PCR performance was investigated by means of qPCR and conventional PCR using PCR fragments of different sizes. DNA fragmentation was analyzed via agarose gel electrophoresis. This study showed that poor PCR amplification was partly caused by inhibition of the DNA polymerase by fragmented DNA from FFPE tissue and not only due to the absence of intact template molecules of sufficient integrity. This PCR inhibition was successfully minimized by increasing the polymerase concentration, dNTP concentration and PCR elongation time thereby allowing for the robust amplification of larger amplicons. This was shown for genomic template DNA as well as for bisulfite-converted template DNA required for DNA methylation analyses. In conclusion, PCR using DNA from FFPE tissue suffers from inhibition which can be alleviated by adaptation of the PCR conditions, therefore allowing for a significant improvement of PCR performance with regard to variability and the generation of larger amplicons. The presented solutions to overcome this PCR inhibition are of tremendous value for clinical chemistry and laboratory medicine.
Conflict of interest statement
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

