AVISPA: a web tool for the prediction and analysis of alternative splicing
- PMID: 24156756
- PMCID: PMC4014802
- DOI: 10.1186/gb-2013-14-10-r114
AVISPA: a web tool for the prediction and analysis of alternative splicing
Abstract
Transcriptome complexity and its relation to numerous diseases underpins the need to predict in silico splice variants and the regulatory elements that affect them. Building upon our recently described splicing code, we developed AVISPA, a Galaxy-based web tool for splicing prediction and analysis. Given an exon and its proximal sequence, the tool predicts whether the exon is alternatively spliced, displays tissue-dependent splicing patterns, and whether it has associated regulatory elements. We assess AVISPA's accuracy on an independent dataset of tissue-dependent exons, and illustrate how the tool can be applied to analyze a gene of interest. AVISPA is available at http://avispa.biociphers.org.
Figures
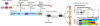
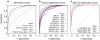

References
-
- Wang ET, Cooper AT. Splicing in disease: disruption of the splicing code and the decoding machinery. Nature. 2007;14:749–761. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

