Bridging the gap between theory and experiment to derive a detailed understanding of hammerhead ribozyme catalysis
- PMID: 24156941
- PMCID: PMC4747252
- DOI: 10.1016/B978-0-12-381286-5.00002-0
Bridging the gap between theory and experiment to derive a detailed understanding of hammerhead ribozyme catalysis
Abstract
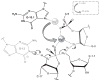
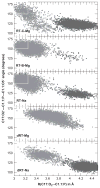
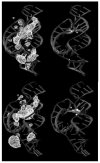
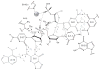
Herein we summarize our progress toward the understanding of hammerhead ribozyme (HHR) catalysis through a multiscale simulation strategy. Simulation results collectively paint a picture of HHR catalysis: HHR first folds to form an electronegative active site pocket to recruit a threshold occupation of cationic charges, either a Mg(2+) ion or multiple monovalent cations. Catalytically active conformations that have good in-line fitness are supported by specific metal ion coordination patterns that involve either a bridging Mg(2+) ion or multiple Na(+) ions, one of which is also in a bridging coordination pattern. In the case of a single Mg(2+) ion bound in the active site, the Mg(2+) ion undergoes a migration that is coupled with deprotonation of the nucleophile (C17:O2'). As the reaction proceeds, the Mg(2+) ion stabilizes the accumulating charge of the leaving group and significantly increases the general acid ability of G8:O2'. Further computational mutagenesis simulations suggest that the disruptions due to mutations may severely impact HHR catalysis at different stages of the reaction. Catalytic mechanisms supported by the simulation results are consistent with available structural and biochemical experiments, and together they advance our understanding of HHR catalysis.
Keywords: RNA; catalysis; combined QM/MM; enzyme; free energy; hammerhead ribozyme; mechanism; simulation.
© 2013 Elsevier Inc. All rights reserved.
Figures

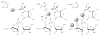

Similar articles
-
Molecular simulations of the pistol ribozyme: unifying the interpretation of experimental data and establishing functional links with the hammerhead ribozyme.RNA. 2019 Nov;25(11):1439-1456. doi: 10.1261/rna.071944.119. Epub 2019 Jul 30. RNA. 2019. PMID: 31363004 Free PMC article.
-
Threshold occupancy and specific cation binding modes in the hammerhead ribozyme active site are required for active conformation.J Mol Biol. 2009 Apr 24;388(1):195-206. doi: 10.1016/j.jmb.2009.02.054. Epub 2009 Mar 2. J Mol Biol. 2009. PMID: 19265710 Free PMC article.
-
Role of Mg2+ in hammerhead ribozyme catalysis from molecular simulation.J Am Chem Soc. 2008 Mar 12;130(10):3053-64. doi: 10.1021/ja076529e. Epub 2008 Feb 14. J Am Chem Soc. 2008. PMID: 18271579 Free PMC article.
-
The hammerhead ribozyme: structure, catalysis, and gene regulation.Prog Mol Biol Transl Sci. 2013;120:1-23. doi: 10.1016/B978-0-12-381286-5.00001-9. Prog Mol Biol Transl Sci. 2013. PMID: 24156940 Free PMC article. Review.
-
Structural Simplicity and Mechanistic Complexity in the Hammerhead Ribozyme.Prog Mol Biol Transl Sci. 2018;159:177-202. doi: 10.1016/bs.pmbts.2018.07.006. Epub 2018 Sep 17. Prog Mol Biol Transl Sci. 2018. PMID: 30340787 Review.
Cited by
-
Force Field for Mg(2+), Mn(2+), Zn(2+), and Cd(2+) Ions That Have Balanced Interactions with Nucleic Acids.J Phys Chem B. 2015 Dec 17;119(50):15460-70. doi: 10.1021/acs.jpcb.5b10423. Epub 2015 Dec 3. J Phys Chem B. 2015. PMID: 26583536 Free PMC article.
-
Dynamical ensemble of the active state and transition state mimic for the RNA-cleaving 8-17 DNAzyme in solution.Nucleic Acids Res. 2019 Nov 4;47(19):10282-10295. doi: 10.1093/nar/gkz773. Nucleic Acids Res. 2019. PMID: 31511899 Free PMC article.
-
Molecular simulations of the pistol ribozyme: unifying the interpretation of experimental data and establishing functional links with the hammerhead ribozyme.RNA. 2019 Nov;25(11):1439-1456. doi: 10.1261/rna.071944.119. Epub 2019 Jul 30. RNA. 2019. PMID: 31363004 Free PMC article.
-
The L-platform/L-scaffold framework: a blueprint for RNA-cleaving nucleic acid enzyme design.RNA. 2020 Feb;26(2):111-125. doi: 10.1261/rna.071894.119. Epub 2019 Nov 27. RNA. 2020. PMID: 31776179 Free PMC article.
-
Multiscale methods for computational RNA enzymology.Methods Enzymol. 2015;553:335-74. doi: 10.1016/bs.mie.2014.10.064. Epub 2015 Jan 22. Methods Enzymol. 2015. PMID: 25726472 Free PMC article.
References
-
- Gilbert W. The RNA, world. Nature. 1986;319:618.
-
- Scott WG. Molecular palaeontology: understanding catalytic mechanisms in the RNA world by excavating clues from a ribozyme three-dimensional structure. Biochem Soc Trans. 1996;24:604–608. - PubMed
-
- Gesteland RF, Cech TR, Atkins JF. The RNA World: The Nature of Modern RNA Suggests a Prebiotic RNA. New York: Cold Spring Harbor Laboratory Press; 1999.
-
- Yarus M. Boundaries for an RNA world. Curr Opin Chem Biol. 1999;3:260–267. - PubMed
-
- Chen X, Li N, Ellington AD. Ribozyme catalysis of metabolism in the RNA world. Chem Biodivers. 2007;4:633–655. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

