Differential regulation of the REGγ-proteasome pathway by p53/TGF-β signalling and mutant p53 in cancer cells
- PMID: 24157709
- PMCID: PMC3876931
- DOI: 10.1038/ncomms3667
Differential regulation of the REGγ-proteasome pathway by p53/TGF-β signalling and mutant p53 in cancer cells
Abstract
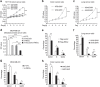
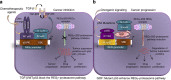
Proteasome activity is frequently enhanced in cancer to accelerate metastasis and tumorigenesis. REGγ, a proteasome activator known to promote p53/p21/p16 degradation, is often overexpressed in cancer cells. Here we show that p53/TGF-β signalling inhibits the REGγ-20S proteasome pathway by repressing REGγ expression. Smad3 and p53 interact on the REGγ promoter via the p53RE/SBE region. Conversely, mutant p53 binds to the REGγ promoter and recruits p300. Importantly, mutant p53 prevents Smad3/N-CoR complex formation on the REGγ promoter, which enhances the activity of the REGγ-20S proteasome pathway and contributes to mutant p53 gain of function. Depletion of REGγ alters the cellular response to p53/TGF-β signalling in drug resistance, proliferation, cell cycle progression and proteasome activity. Moreover, p53 mutations show a positive correlation with REGγ expression in cancer samples. These findings suggest that targeting REGγ-20S proteasome for cancer therapy may be applicable to human tumours with abnormal p53/Smad protein status. Furthermore, this study demonstrates a link between p53/TGF-β signalling and the REGγ-20S proteasome pathway, and provides insight into the REGγ/p53 feedback loop.
Figures








References
-
- Ma C. P., Slaughter C. A. & DeMartino G. N. Identification, purification, and characterization of a protein activator (PA28) of the 20 S proteasome (macropain). J. Biol. Chem. 267, 10515–10523 (1992). - PubMed
-
- Dubiel W., Pratt G., Ferrell K. & Rechsteiner M. Purification of an 11 S regulator of the multicatalytic protease. J. Biol. Chem. 267, 22369–22377 (1992). - PubMed
-
- Li X. et al.. The SRC-3/AIB1 coactivator is degraded in a ubiquitin- and ATP-independent manner by the REGgamma proteasome. Cell 124, 381–392 (2006). - PubMed
-
- Li X. et al.. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Mol. Cell 26, 831–842 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

