High-resolution mapping of the spatial organization of a bacterial chromosome
- PMID: 24158908
- PMCID: PMC3927313
- DOI: 10.1126/science.1242059
High-resolution mapping of the spatial organization of a bacterial chromosome
Abstract
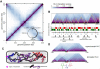
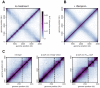
Chromosomes must be highly compacted and organized within cells, but how this is achieved in vivo remains poorly understood. We report the use of chromosome conformation capture coupled with deep sequencing (Hi-C) to map the structure of bacterial chromosomes. Analysis of Hi-C data and polymer modeling indicates that the Caulobacter crescentus chromosome consists of multiple, largely independent spatial domains that are probably composed of supercoiled plectonemes arrayed into a bottle brush-like fiber. These domains are stable throughout the cell cycle and are reestablished concomitantly with DNA replication. We provide evidence that domain boundaries are established by highly expressed genes and the formation of plectoneme-free regions, whereas the histone-like protein HU and SMC (structural maintenance of chromosomes) promote short-range compaction and the colinearity of chromosomal arms, respectively. Collectively, our results reveal general principles for the organization and structure of chromosomes in vivo.
Figures


References
-
- Booker BM, Deng S, Higgins NP. Mol Microbiol. 2010;78:1348. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

