A bioinformatics approach identifies signal transducer and activator of transcription-3 and checkpoint kinase 1 as upstream regulators of kidney injury molecule-1 after kidney injury
- PMID: 24158981
- PMCID: PMC3871775
- DOI: 10.1681/ASN.2013020161
A bioinformatics approach identifies signal transducer and activator of transcription-3 and checkpoint kinase 1 as upstream regulators of kidney injury molecule-1 after kidney injury
Abstract
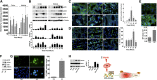
Kidney injury molecule-1 (KIM-1)/T cell Ig and mucin domain-containing protein-1 (TIM-1) is upregulated more than other proteins after AKI, and it is highly expressed in renal damage of various etiologies. In this capacity, KIM-1/TIM-1 acts as a phosphatidylserine receptor on the surface of injured proximal tubular epithelial cells, mediating phagocytosis of apoptotic cells, and it may also act as a costimulatory molecule for immune cells. Despite recognition of KIM-1 as an important therapeutic target for kidney disease, the regulators of KIM-1 transcription in the kidney remain unknown. Using a bioinformatics approach, we identified upstream regulators of KIM-1 after AKI. In response to tubular injury in rat and human kidneys or oxidant stress in human proximal tubular epithelial cells (HPTECs), KIM-1 expression increased significantly in a manner that corresponded temporally and regionally with increased phosphorylation of checkpoint kinase 1 (Chk1) and STAT3. Both ischemic and oxidant stress resulted in a dramatic increase in reactive oxygen species that phosphorylated and activated Chk1, which subsequently bound to STAT3, phosphorylating it at S727. Furthermore, STAT3 bound to the KIM-1 promoter after ischemic and oxidant stress, and pharmacological or genetic induction of STAT3 in HPTECs increased KIM-1 mRNA and protein levels. Conversely, inhibition of STAT3 using siRNAs or dominant negative mutants reduced KIM-1 expression in a kidney cancer cell line (769-P) that expresses high basal levels of KIM-1. These observations highlight Chk1 and STAT3 as critical upstream regulators of KIM-1 expression after AKI and may suggest novel approaches for therapeutic intervention.
Figures




References
-
- Ichimura T, Bonventre JV, Bailly V, Wei H, Hession CA, Cate RL, Sanicola M: Kidney injury molecule-1 (KIM-1), a putative epithelial cell adhesion molecule containing a novel immunoglobulin domain, is up-regulated in renal cells after injury. J Biol Chem 273: 4135–4142, 1998 - PubMed
-
- Amin RP, Vickers AE, Sistare F, Thompson KL, Roman RJ, Lawton M, Kramer J, Hamadeh HK, Collins J, Grissom S, Bennett L, Tucker CJ, Wild S, Kind C, Oreffo V, Davis JW, 2nd, Curtiss S, Naciff JM, Cunningham M, Tennant R, Stevens J, Car B, Bertram TA, Afshari CA: Identification of putative gene based markers of renal toxicity. Environ Health Perspect 112: 465–479, 2004 - PMC - PubMed
-
- Vaidya VS, Ozer JS, Dieterle F, Collings FB, Ramirez V, Troth S, Muniappa N, Thudium D, Gerhold D, Holder DJ, Bobadilla NA, Marrer E, Perentes E, Cordier A, Vonderscher J, Maurer G, Goering PL, Sistare FD, Bonventre JV: Kidney injury molecule-1 outperforms traditional biomarkers of kidney injury in preclinical biomarker qualification studies. Nat Biotechnol 28: 478–485, 2010 - PMC - PubMed
-
- Vaidya VS, Ramirez V, Ichimura T, Bobadilla NA, Bonventre JV: Urinary kidney injury molecule-1: A sensitive quantitative biomarker for early detection of kidney tubular injury. Am J Physiol Renal Physiol 290: F517–F529, 2006 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

