Structural insights into Resveratrol's antagonist and partial agonist actions on estrogen receptor alpha
- PMID: 24160181
- PMCID: PMC4015837
- DOI: 10.1186/1472-6807-13-27
Structural insights into Resveratrol's antagonist and partial agonist actions on estrogen receptor alpha
Abstract
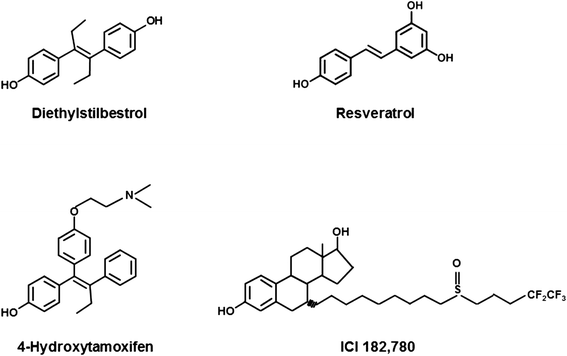
Background: Resveratrol, a naturally occurring stilbene, has been categorized as a phytoestrogen due to its ability to compete with natural estrogens for binding to estrogen receptor alpha (ERα) and modulate the biological responses exerted by the receptor. Biological effects of resveratrol (RES) on estrogen receptor alpha (ERα) remain highly controversial, since both estrogenic and anti-estrogenic properties were observed.
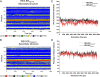
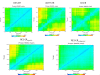
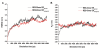
Results: Here, we provide insight into the structural basis of the agonist/antagonist effects of RES on ERα ligand binding domain (LBD). Using atomistic simulation, we found that RES bound ERα monomer in antagonist conformation, where Helix 12 moves away from the ligand pocket and orients into the co-activator binding groove of LBD, is more stable than RES bound ERα in agonist conformation, where Helix 12 lays over the ligand binding pocket. Upon dimerization, the agonistic conformation of RES-ERα dimer becomes more stable compared to the corresponding monomer but still remains less stable compared to the corresponding dimer in antagonist conformation. Interestingly, while the binding pocket and the binding contacts of RES to ERα are similar to those of pure agonist diethylstilbestrol (DES), the binding energy is much less and the hydrogen bonding contacts also differ providing clues for the partial agonistic character of RES on ERα.
Conclusions: Our Molecular Dynamics simulation of RES-ERα structures with agonist and antagonist orientations of Helix 12 suggests RES action is more similar to Selective Estrogen Receptor Modulator (SERM) opening up the importance of cellular environment and active roles of co-regulator proteins in a given system. Our study reveals that potential co-activators must compete with the Helix 12 and displace it away from the activator binding groove to enhance the agonistic activity.
Figures



References
-
- Siemann EH, Creasy LL. Concentration of the phytoalexin resveratrol in wine. Am J Enol Vitic. 1992;13:49–52.
-
- Frankel EN, Waterhouse AL, Kinsella JE. Inhibition of human LDL oxidation by resveratrol. Lancet. 1993;13:1103–1104. - PubMed
-
- Pace-Asciak CR, Hahn S, Diamandis EP, Soleas G, Goldberg DM. The red wine phenolics trans-resveratrol and quercetin block human platelet aggregation and eicosanoid synthesis: implications for protection against coronary heart disease. Clin Chim Acta. 1995;13:207–219. doi: 10.1016/0009-8981(95)06045-1. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

