Standardization of whole blood immune phenotype monitoring for clinical trials: panels and methods from the ONE study
- PMID: 24160259
- PMCID: PMC3827923
- DOI: 10.1186/2047-1440-2-17
Standardization of whole blood immune phenotype monitoring for clinical trials: panels and methods from the ONE study
Abstract
Background: Immune monitoring by flow cytometry is a fast and highly informative way of studying the effects of novel therapeutics aimed at reducing transplant rejection or treating autoimmune diseases. The ONE Study consortium has recently initiated a series of clinical trials aimed at using different cell therapies to promote tolerance to renal allografts. To compare the effectiveness of different cell therapies, the consortium developed a robust immune monitoring strategy, including procedures for whole blood (WB) leukocyte subset profiling by flow cytometry.
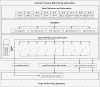
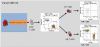
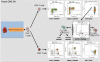
Methods: Six leukocyte profiling panels computing 7- to 9-surface marker antigens for monitoring the major leukocyte subsets as well as characteristics of T cell, B cell, and dendritic cell (DC) subsets were designed. The precision and variability of these panels were estimated. The assay was standardized within eight international laboratories using Flow-Set Pro beads for mean fluorescence intensity target definition and the flow cytometer setup procedure. Standardization was demonstrated by performing inter-site comparisons.
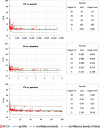
Results: Optimized methods for sample collection, storage, preparation, and analysis were established, including protocols for gating target subsets. WB specimen age testing demonstrated that staining must be performed within 4 hours of sample collection to keep variability low, meaning less than or equal to 10% for the majority of defined leukocyte subsets. Inter-site comparisons between all participating centers testing shipped normal WB revealed good precision, with a variability of 0.05% to 30% between sites. Intra-assay analyses revealed a variability of 0.05% to 20% for the majority of subpopulations. This was dependent on the frequency of the particular subset, with smaller subsets showing higher variability. The intra-assay variability performance defined limits of quantitation (LoQ) for subsets, which will be the basis for assessing statistically significant differences achieved by the different cell therapies.
Conclusions: Local performance and central analysis of the ONE Study flow cytometry panel yields acceptable variability in a standardized assay at multiple international sites. These panels and procedures with WB allow unmanipulated analysis of changes in absolute cell numbers of leukocyte subsets in single- or multicenter clinical trials. Accordingly, we propose the ONE Study panel may be adopted as a standardized method for monitoring patients in clinical trials enrolling transplant patients, particularly trials of novel tolerance promoting therapies, to facilitate fair and meaningful comparisons between trials.
Figures





References
-
- Broichhausen C, Riquelme P, Geissler EK, Hutchinson JA. Regulatory macrophages as therapeutic targets and therapeutic agents in solid organ transplantation. Curr Opin Organ Trans. 2012;17(4):332–342. - PubMed
-
- Brunstein CG, Miller JS, Cao Q, McKenna DH, Hippen KL, Curtsinger J, Defor T, Levine BL, June CH, Rubinstein P, McGlave PB, Blazar BR, Wagner JE. Infusion of ex vivo expanded T regulatory cells in adults transplanted with umbilical cord blood: safety profile and detection kinetics. Blood. 2011;117(3):1061–1070. doi: 10.1182/blood-2010-07-293795. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

