The distribution of word matches between Markovian sequences with periodic boundary conditions
- PMID: 24160839
- PMCID: PMC3880068
- DOI: 10.1089/cmb.2012.0277
The distribution of word matches between Markovian sequences with periodic boundary conditions
Abstract
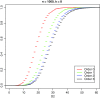
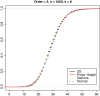
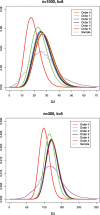
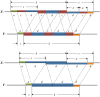
Word match counts have traditionally been proposed as an alignment-free measure of similarity for biological sequences. The D(2) statistic, which simply counts the number of exact word matches between two sequences, is a useful test bed for developing rigorous mathematical results, which can then be extended to more biologically useful measures. The distributional properties of the D(2) statistic under the null hypothesis of identically and independently distributed letters have been studied extensively, but no comprehensive study of the D(2) distribution for biologically more realistic higher-order Markovian sequences exists. Here we derive exact formulas for the mean and variance of the D(2) statistic for Markovian sequences of any order, and demonstrate through Monte Carlo simulations that the entire distribution is accurately characterized by a Pólya-Aeppli distribution for sequence lengths of biological interest. The approach is novel in that Markovian dependency is defined for sequences with periodic boundary conditions, and this enables exact analytic formulas for the mean and variance to be derived. We also carry out a preliminary comparison between the approximate D(2) distribution computed with the theoretical mean and variance under a Markovian hypothesis and an empirical D(2) distribution from the human genome.
Figures




References
-
- Burden C.J., Kantorovitz M.R., and Wilson S.R.2008. Approximate word matches between two random sequences. Ann. Appl. Probab. 18, 1–21
-
- Burden C.J., Jing J., Forêt S., and Wilson S.R.2012. Application of k-word match statistics to the clustering of proteins with repeated domains. In Colubi A., Fokianos K., Kontoghiorghes E., and González-Rodríguez G., eds. Proceedings of COMPSTAT 2012, 20th International Conference on Computational Statistics 131–142
-
- Burden C.J., Jing J., and Wilson S.R.2012. Alignment-free sequence comparison for biologically realistic sequences of moderate length. Stat. Appl. Genet. Mol. Biol. 11, Article 3. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

