Comprehensive profiling of Epstein-Barr virus-encoded miRNA species associated with specific latency types in tumor cells
- PMID: 24161012
- PMCID: PMC4231337
- DOI: 10.1186/1743-422X-10-314
Comprehensive profiling of Epstein-Barr virus-encoded miRNA species associated with specific latency types in tumor cells
Abstract
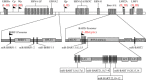
Background: Epstein-Barr virus (EBV) is an etiological cause of many human lymphocytic and epithelial malignancies. EBV expresses different genes that are associated with three latency types. To date, as many as 44 EBV-encoded miRNA species have been found, but their comprehensive profiles in the three types of latent infection that are associated with various types of tumors are not well documented.
Methods: In the present study, we utilized poly (A)-tailed quantitative real-time RT-PCR in combination with microarray analysis to measure the relative abundances of viral miRNA species in a subset of representative lymphoid and epithelial tumor cells with various EBV latency types.
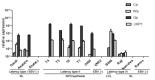
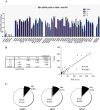
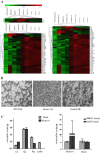
Results: Our findings showed that the miR-BHRF1 and miR-BART families were expressed differentially in a tissue- and latency type-dependent manner. Specifically, in nasopharyngeal carcinoma (NPC) tissues and the EBV-positive cell line C666-1, the miR-BART family accounted for more than 10% of all detected miRNAs, suggesting that these miRNAs have important roles in maintaining latent EBV infections and in driving NPC tumorigenesis. In addition, EBV miRNA-based clustering analysis clearly distinguished between the three distinct EBV latency types, and our results suggested that a switch from type I to type III latency might occur in the Daudi BL cell line.
Conclusions: Our data provide a comprehensive profiling of the EBV miRNA transcriptome that is associated with specific tumor cells in the three types of latent EBV infection states. EBV miRNA species represent a cluster of non-encoding latency biomarkers that are differentially expressed in tumor cells and may help to distinguish between the different latency types.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

