CRISPRs: molecular signatures used for pathogen subtyping
- PMID: 24162568
- PMCID: PMC3911090
- DOI: 10.1128/AEM.02790-13
CRISPRs: molecular signatures used for pathogen subtyping
Abstract
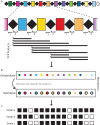
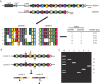
Rapid and accurate strain identification is paramount in the battle against microbial outbreaks, and several subtyping approaches have been developed. One such method uses clustered regular interspaced short palindromic repeats (CRISPRs), DNA repeat elements that are present in approximately half of all bacteria. Though their signature function is as an adaptive immune system against invading DNA such as bacteriophages and plasmids, CRISPRs also provide an excellent framework for pathogen tracking and evolutionary studies. Analysis of the spacer DNA sequences that reside between the repeats has been tremendously useful for bacterial subtyping during molecular epidemiological investigations. Subtyping, or strain identification, using CRISPRs has been employed in diverse Gram-positive and Gram-negative bacteria, including Mycobacterium tuberculosis, Salmonella enterica, and the plant pathogen Erwinia amylovora. This review discusses the several ways in which CRISPR sequences are exploited for subtyping. This includes the well-established spoligotyping methodologies that have been used for 2 decades to type Mycobacterium species, as well as in-depth consideration of newer, higher-throughput CRISPR-based protocols.
Figures



Similar articles
-
Erwinia amylovora CRISPR elements provide new tools for evaluating strain diversity and for microbial source tracking.PLoS One. 2012;7(7):e41706. doi: 10.1371/journal.pone.0041706. Epub 2012 Jul 31. PLoS One. 2012. PMID: 22860008 Free PMC article.
-
New clustered regularly interspaced short palindromic repeat locus spacer pair typing method based on the newly incorporated spacer for Salmonella enterica.J Clin Microbiol. 2014 Aug;52(8):2955-62. doi: 10.1128/JCM.00696-14. Epub 2014 Jun 4. J Clin Microbiol. 2014. PMID: 24899040 Free PMC article.
-
Clustered regularly interspaced short palindromic repeats (CRISPRs) analysis of members of the Mycobacterium tuberculosis complex.Methods Mol Biol. 2015;1247:373-89. doi: 10.1007/978-1-4939-2004-4_27. Methods Mol Biol. 2015. PMID: 25399110
-
Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes.Biol Chem. 2011 Apr;392(4):277-89. doi: 10.1515/BC.2011.042. Epub 2011 Feb 7. Biol Chem. 2011. PMID: 21294681 Review.
-
[Clustered regularly interspaced short palindromic repeats: structure, function and application--a review].Wei Sheng Wu Xue Bao. 2008 Nov;48(11):1549-55. Wei Sheng Wu Xue Bao. 2008. PMID: 19149174 Review. Chinese.
Cited by
-
Source Tracking Based on Core Genome SNV and CRISPR Typing of Salmonella enterica Serovar Heidelberg Isolates Involved in Foodborne Outbreaks in Québec, 2012.Front Microbiol. 2020 Jun 17;11:1317. doi: 10.3389/fmicb.2020.01317. eCollection 2020. Front Microbiol. 2020. PMID: 32625190 Free PMC article.
-
Comparative Genomic Analysis of Vibrio cincinnatiensis Provides Insights into Genetic Diversity, Evolutionary Dynamics, and Pathogenic Traits of the Species.Int J Mol Sci. 2022 Apr 20;23(9):4520. doi: 10.3390/ijms23094520. Int J Mol Sci. 2022. PMID: 35562911 Free PMC article.
-
CRISPR elements provide a new framework for the genealogy of the citrus canker pathogen Xanthomonas citri pv. citri.BMC Genomics. 2019 Dec 2;20(1):917. doi: 10.1186/s12864-019-6267-z. BMC Genomics. 2019. PMID: 31791238 Free PMC article.
-
CRISPR-cas subtype I-Fb in Acinetobacter baumannii: evolution and utilization for strain subtyping.PLoS One. 2015 Feb 23;10(2):e0118205. doi: 10.1371/journal.pone.0118205. eCollection 2015. PLoS One. 2015. PMID: 25706932 Free PMC article.
-
Cronobacter sakazakii, Cronobacter malonaticus, and Cronobacter dublinensis Genotyping Based on CRISPR Locus Diversity.Front Microbiol. 2019 Aug 28;10:1989. doi: 10.3389/fmicb.2019.01989. eCollection 2019. Front Microbiol. 2019. PMID: 31555228 Free PMC article.
References
-
- Sabat A, Budimir A, Nashev D, Sá-Leão R, van Dijl J, Laurent F, Grundmann H, Friedrich AW; ESCMID Study Group of Epidemiological Markers (ESGEM) 2013. Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Euro Surveill. 18:20380. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

