Protein O-glucosylation in Lactobacillus buchneri
- PMID: 24162649
- PMCID: PMC4396861
- DOI: 10.1007/s10719-013-9505-7
Protein O-glucosylation in Lactobacillus buchneri
Abstract
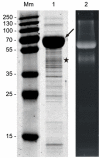
Based on the previous demonstration of surface (S-) layer protein glycosylation in Lactobacillus buchneri 41021/251 and because of general advantages of lactic acid bacteria for applied research, protein glycosylation in this bacterial species was investigated in detail. The cell surface of L. buchneri CD034 is completely covered with an oblique 2D crystalline array (lattice parameters, a = 5.9 nm; b = 6.2 nm; γ ~ 77°) formed by self-assembly of the S-layer protein SlpB. Biochemical and mass spectrometric analyses revealed that SlpB is the most abundant protein and that it is O-glycosylated at four serine residues within the sequence S(152)-A-S(154)-S(155)-A-S(157) with, on average, seven Glc(α1-6) residues, each. Subcellular fractionation of strain CD034 indicated a sequential order of SlpB export and glucosylation as evidenced by lack of glucosylation of cytosolic SlpB. Protein glycosylation analysis was extended to strain L. buchneri NRRL B-30929 where an analogous glucosylation scenario could be detected, with the S-layer glycoprotein SlpN containing an O-glycosylation motif identical to that of SlpB. This corroborates previous data on S-layer protein glucosylation of strain 41021/251 and let us propose a species-wide S-layer protein O-glucosylation in L. buchneri targeted at the sequence motif S-A-S-S-A-S. Search of the L. buchneri genomes for the said glucosylation motif revealed one further ORF, encoding the putative glycosyl-hydrolase LbGH25B and LbGH25N in L. buchneri CD034 and NRRL B-30929, respectively, for which we have indications of a glycosylation comparable to that of the S-layer proteins. These findings demonstrate the presence of a distinct protein O-glucosylation system in Gram-positive and beneficial microbes.
Figures




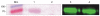


References
-
- Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. Essentials of glycobiology. 2nd edn. Cold Spring Harbor Laboratory Press; Cold Spring Habor: 2009. p. 484. - PubMed
-
- Upreti RK, Kumar M, Shankar V. Bacterial glycoproteins: functions, biosynthesis and applications. Proteomics. 2003;3:363–379. - PubMed
-
- Schmidt MA, Riley LW, Benz I. Sweet new world: glycoproteins in bacterial pathogens. Trends Microbiol. 2003;11:554–561. - PubMed
-
- Hitchen PG, Dell A. Bacterial glycoproteomics. Microbiology. 2006;152:1575–1580. - PubMed
-
- Nothaft H, Szymanski CM. Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 2010;8:765–778. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

