A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA
- PMID: 24162923
- PMCID: PMC3852148
- DOI: 10.1038/nmeth.2701
A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA
Abstract
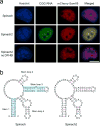
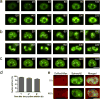
Imaging RNA in living cells is a challenging problem in cell biology. One strategy for genetically encoding fluorescent RNAs is to express them as fusions with Spinach, an 'RNA mimic of GFP'. We found that Spinach was dimmer than expected when used to tag constructs in living cells owing to a combination of thermal instability and a propensity for misfolding. Using systematic mutagenesis, we generated Spinach2 that overcomes these issues and can be used to image diverse RNAs. Using Spinach2, we detailed the dynamics of the CGG trinucleotide repeat-containing 'toxic RNA' associated with Fragile X-associated tremor/ataxia syndrome, and show that these RNAs form nuclear foci with unexpected morphological plasticity that is regulated by the cell cycle and by small molecules. Together, these data demonstrate that Spinach2 exhibits improved versatility for fluorescently labeling RNAs in living cells.
Figures
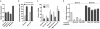



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

