REST mediates androgen receptor actions on gene repression and predicts early recurrence of prostate cancer
- PMID: 24163104
- PMCID: PMC3902919
- DOI: 10.1093/nar/gkt921
REST mediates androgen receptor actions on gene repression and predicts early recurrence of prostate cancer
Abstract
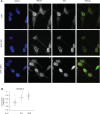
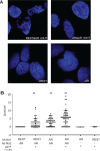
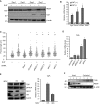
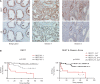
The androgen receptor (AR) is a key regulator of prostate tumorgenesis through actions that are not fully understood. We identified the repressor element (RE)-1 silencing transcription factor (REST) as a mediator of AR actions on gene repression. Chromatin immunoprecipitation showed that AR binds chromatin regions containing well-characterized cis-elements known to mediate REST transcriptional repression, while cell imaging studies confirmed that REST and AR closely co-localize in vivo. Androgen-induced gene repression also involves modulation of REST protein turnover through actions on the ubiquitin ligase β-TRCP. Androgen deprivation or AR blockage with inhibitor MDV3100 (Enzalutamide) leads to neuroendocrine (NE) differentiation, a phenomenon that is mimicked by REST inactivation. Gene expression profiling revealed that REST not only acts to repress neuronal genes but also genes involved in cell cycle progression, including Aurora Kinase A, that has previously been implicated in the growth of NE-like castration-resistant tumors. The analysis of prostate cancer tissue microarrays revealed that tumors with reduced expression of REST have higher probability of early recurrence, independently of their Gleason score. The demonstration that REST modulates AR actions in prostate epithelia and that REST expression is negatively correlated with disease recurrence after prostatectomy, invite a deeper characterization of its role in prostate carcinogenesis.
Figures




References
-
- Epstein JI, Amin M, Boccon-Gibod L, Egevad L, Humphrey PA, Mikuz G, Newling D, Nilsson S, Sakr W, Srigley JR, et al. Prognostic factors and reporting of prostate carcinoma in radical prostatectomy and pelvic lymphadenectomy specimens. Scand. J. Urol. Nephrol. 2005:34–63. - PubMed
-
- Pienta KJ, Bradley D. Mechanisms underlying the development of androgen-independent prostate cancer. Clin. Cancer Res. 2006;12:1665–1671. - PubMed
-
- Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr. Rev. 2004;25:276–308. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

