RADAR: a rigorously annotated database of A-to-I RNA editing
- PMID: 24163250
- PMCID: PMC3965033
- DOI: 10.1093/nar/gkt996
RADAR: a rigorously annotated database of A-to-I RNA editing
Abstract
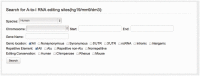
We present RADAR--a rigorously annotated database of A-to-I RNA editing (available at http://RNAedit.com). The identification of A-to-I RNA editing sites has been dramatically accelerated in the past few years by high-throughput RNA sequencing studies. RADAR includes a comprehensive collection of A-to-I RNA editing sites identified in humans (Homo sapiens), mice (Mus musculus) and flies (Drosophila melanogaster), together with extensive manually curated annotations for each editing site. RADAR also includes an expandable listing of tissue-specific editing levels for each editing site, which will facilitate the assignment of biological functions to specific editing sites.
Figures

References
-
- Barbon A, Barlati S. Genomic organization, proposed alternative splicing mechanisms, and RNA editing structure of GRIK1. Cytogenet. Cell Genet. 2000;88:236–239. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

