GTP dysregulation in Bacillus subtilis cells lacking (p)ppGpp results in phenotypic amino acid auxotrophy and failure to adapt to nutrient downshift and regulate biosynthesis genes
- PMID: 24163341
- PMCID: PMC3911124
- DOI: 10.1128/JB.00918-13
GTP dysregulation in Bacillus subtilis cells lacking (p)ppGpp results in phenotypic amino acid auxotrophy and failure to adapt to nutrient downshift and regulate biosynthesis genes
Abstract
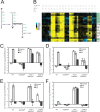
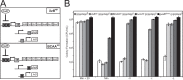
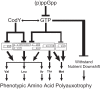
The nucleotide (p)ppGpp inhibits GTP biosynthesis in the Gram-positive bacterium Bacillus subtilis. Here we examined how this regulation allows cells to grow in the absence of amino acids. We showed that B. subtilis cells lacking (p)ppGpp, due to either deletions or point mutations in all three (p)ppGpp synthetase genes, yjbM, ywaC, and relA, strongly require supplementation of leucine, isoleucine, valine, methionine, and threonine and modestly require three additional amino acids. This polyauxotrophy is rescued by reducing GTP levels. Reduction of GTP levels activates transcription of genes responsible for the biosynthesis of the five strongly required amino acids by inactivating the transcription factor CodY, which represses the ybgE, ilvD, ilvBHC-leuABCD, ilvA, ywaA, and hom-thrCB operons, and by a CodY-independent activation of transcription of the ilvA, ywaA, hom-thrCB, and metE operons. Interestingly, providing the eight required amino acids does not allow for colony formation of (p)ppGpp(0) cells when transitioning from amino acid-replete medium to amino acid-limiting medium, and we found that this is due to an additional role that (p)ppGpp plays in protecting cells during nutrient downshifts. We conclude that (p)ppGpp allows adaptation to amino acid limitation by a combined effect of preventing death during metabolic transitions and sustaining growth by activating amino acid biosynthesis. This ability of (p)ppGpp to integrate a general stress response with a targeted reprogramming of gene regulation allows appropriate adaptation and is likely conserved among diverse bacteria.
Figures




References
-
- Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980–5990 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

