Phenotypic and genotypic convergences are influenced by historical contingency and environment in yeast
- PMID: 24164389
- PMCID: PMC4439200
- DOI: 10.1111/evo.12302
Phenotypic and genotypic convergences are influenced by historical contingency and environment in yeast
Abstract
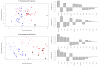
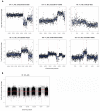
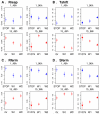
Different organisms have independently and recurrently evolved similar phenotypic traits at different points throughout history. This phenotypic convergence may be caused by genotypic convergence and in addition, constrained by historical contingency. To investigate how convergence may be driven by selection in a particular environment and constrained by history, we analyzed nine life-history traits and four metabolic traits during an experimental evolution of six yeast strains in four different environments. In each of the environments, the population converged toward a different multivariate phenotype. However, the evolution of most traits, including fitness components, was constrained by history. Phenotypic convergence was partly associated with the selection of mutations in genes involved in the same pathway. By further investigating the convergence in one gene, BMH1, mutated in 20% of the evolved populations, we show that both the history and the environment influenced the types of mutations (missense/nonsense), their location within the gene itself, as well as their effects on multiple traits. However, these effects could not be easily predicted from ancestors' phylogeny or past selection. Combined, our data highlight the role of pleiotropy and epistasis in shaping a rugged fitness landscape.
Keywords: 14-3-3 protein; Adaptive landscape; experimental evolution; life-history evolution; pleiotropy.
© 2013 The Author(s). Evolution © 2013 The Society for the Study of Evolution.
Figures

References
-
- Aitken A. 14-3-3 proteins: A historic overview. Seminars in Cancer Biology. 2006;16:162–172. - PubMed
-
- Arendt J, Reznick D. Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends in Ecology & Evolution. 2008;23:26–32. - PubMed
-
- Arnold SJ, Pfrender ME, Jones AG. The adaptive landscape as a conceptual bridge between micro-and macroevolution. Genetica. 2001;112:9–32. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
