Structural models of ligand-gated ion channels: sites of action for anesthetics and ethanol
- PMID: 24164436
- PMCID: PMC3959612
- DOI: 10.1111/acer.12283
Structural models of ligand-gated ion channels: sites of action for anesthetics and ethanol
Abstract
The molecular mechanism(s) of action of anesthetic, and especially, intoxicating doses of alcohol (ethanol [EtOH]) have been of interest even before the advent of the Research Society on Alcoholism. Recent physiological, genetic, and biochemical studies have pin-pointed molecular targets for anesthetics and EtOH in the brain as ligand-gated ion channel (LGIC) membrane proteins, especially the pentameric (5 subunit) Cys-loop superfamily of neurotransmitter receptors including nicotinic acetylcholine (nAChRs), GABAA (GABAA Rs), and glycine receptors (GlyRs). The ability to demonstrate molecular and structural elements of these proteins critical for the behavioral effects of these drugs on animals and humans provides convincing evidence for their role in the drugs' actions. Amino acid residues necessary for pharmacologically relevant allosteric modulation of LGIC function by anesthetics and EtOH have been identified in these channel proteins. Site-directed mutagenesis revealed potential allosteric modulatory sites in both the trans-membrane domain (TMD) and extracellular domain (ECD). Potential sites of action and binding have been deduced from homology modeling of other LGICs with structures known from crystallography and cryo-electron microscopy studies. Direct information about ligand binding in the TMD has been obtained by photoaffinity labeling, especially in GABAA Rs. Recent structural information from crystallized procaryotic (ELIC and GLIC) and eukaryotic (GluCl) LGICs allows refinement of the structural models including evaluation of possible sites of EtOH action.
Keywords: ELIC; Ethanol Sites of Action; GABAA Receptors; GLIC; GluCl Pentameric Ion Channels; Loop 2.
Copyright © 2013 by the Research Society on Alcoholism.
Figures
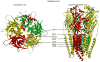


References
-
- Arevalo E, Chiara DC, Forman SA, Cohen JB, Miller KW. Gating-enhanced accessibility of hydrophobic sites within the transmembrane region of the nicotinic acetylcholine receptor’s {delta}-subunit. A time-resolved photolabeling study. J Biol Chem. 2005;280:13631–13640. - PubMed
-
- Bali M, Akabas MH. Defining the propofol binding site location on the GABAA receptor. Mol Pharmacol. 2004;65:68–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

