MoeH5: a natural glycorandomizer from the moenomycin biosynthetic pathway
- PMID: 24164498
- PMCID: PMC3961715
- DOI: 10.1111/mmi.12437
MoeH5: a natural glycorandomizer from the moenomycin biosynthetic pathway
Abstract
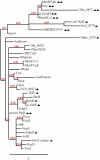
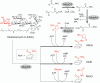
The biosynthesis of the phosphoglycolipid antibiotic moenomycin A attracts the attention of researchers hoping to develop new moenomycin-based antibiotics against multidrug resistant Gram-positive infections. There is detailed understanding of most steps of this biosynthetic pathway in Streptomyces ghanaensis (ATCC14672), except for the ultimate stage, where a single pentasaccharide intermediate is converted into a set of unusually modified final products. Here we report that only one gene, moeH5, encoding a homologue of the glutamine amidotransferase (GAT) enzyme superfamily, is responsible for the observed diversity of terminally decorated moenomycins. Genetic and biochemical evidence support the idea that MoeH5 is a novel member of the GAT superfamily, whose homologues are involved in the synthesis of various secondary metabolites as well as K and O antigens of bacterial lipopolysaccharide. Our results provide insights into the mechanism of MoeH5 and its counterparts, and give us a new tool for the diversification of phosphoglycolipid antibiotics.
© 2013 John Wiley & Sons Ltd.
Figures





Similar articles
-
A gene cluster for the biosynthesis of moenomycin family antibiotics in the genome of teicoplanin producer Actinoplanes teichomyceticus.Appl Microbiol Biotechnol. 2016 Sep;100(17):7629-38. doi: 10.1007/s00253-016-7685-3. Epub 2016 Jun 25. Appl Microbiol Biotechnol. 2016. PMID: 27344593
-
Complete characterization of the seventeen step moenomycin biosynthetic pathway.Biochemistry. 2009 Sep 22;48(37):8830-41. doi: 10.1021/bi901018q. Biochemistry. 2009. PMID: 19640006 Free PMC article.
-
A streamlined metabolic pathway for the biosynthesis of moenomycin A.Chem Biol. 2007 Mar;14(3):257-67. doi: 10.1016/j.chembiol.2007.01.008. Chem Biol. 2007. PMID: 17379141 Free PMC article.
-
The molecular biology of moenomycins: towards novel antibiotics based on inhibition of bacterial peptidoglycan glycosyltransferases.Biol Chem. 2010 May;391(5):499-504. doi: 10.1515/BC.2010.053. Biol Chem. 2010. PMID: 20302515 Review.
-
Nature's combinatorial biosynthesis and recently engineered production of nucleoside antibiotics in Streptomyces.World J Microbiol Biotechnol. 2017 Apr;33(4):66. doi: 10.1007/s11274-017-2233-6. Epub 2017 Mar 4. World J Microbiol Biotechnol. 2017. PMID: 28260195 Review.
Cited by
-
Genetic approaches to improve clorobiocin production in Streptomyces roseochromogenes NRRL 3504.Appl Microbiol Biotechnol. 2022 Feb;106(4):1543-1556. doi: 10.1007/s00253-022-11814-4. Epub 2022 Feb 11. Appl Microbiol Biotechnol. 2022. PMID: 35147743 Free PMC article.
-
Prospects for novel inhibitors of peptidoglycan transglycosylases.Bioorg Chem. 2014 Aug;55(100):16-26. doi: 10.1016/j.bioorg.2014.05.007. Epub 2014 May 21. Bioorg Chem. 2014. PMID: 24924926 Free PMC article. Review.
-
Structural diversity, bioactivity, and biosynthesis of phosphoglycolipid family antibiotics: Recent advances.BBA Adv. 2022 Nov 17;2:100065. doi: 10.1016/j.bbadva.2022.100065. eCollection 2022. BBA Adv. 2022. PMID: 37082588 Free PMC article.
-
Genetic Engineering of Streptomyces ghanaensis ATCC14672 for Improved Production of Moenomycins.Microorganisms. 2021 Dec 24;10(1):30. doi: 10.3390/microorganisms10010030. Microorganisms. 2021. PMID: 35056478 Free PMC article.
-
Genome Engineering Approaches to Improve Nosokomycin A Production by Streptomyces ghanaensis B38.3.Indian J Microbiol. 2019 Mar;59(1):109-111. doi: 10.1007/s12088-018-0761-x. Epub 2018 Sep 25. Indian J Microbiol. 2019. PMID: 30728639 Free PMC article.
References
-
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55:539 – 552. - PubMed
-
- Belanger M, Burrows LL, Lam JS. Functional analysis of genes responsible for the synthesis of the B-band O antigen of Pseudomonas aeruginosa serotype O6 lipopolysaccharide. Microbiology. 1999;145:3505–3521. - PubMed
-
- Campbell J, Singh AK, Santa Maria JP, Jr., Kim Y, Brown S, Swoboda JG, Mylonakis E, Wilkinson BJ, Walker S. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem Biol. 2011;6:106–116. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

