Copy number alterations in small intestinal neuroendocrine tumors determined by array comparative genomic hybridization
- PMID: 24165089
- PMCID: PMC3819709
- DOI: 10.1186/1471-2407-13-505
Copy number alterations in small intestinal neuroendocrine tumors determined by array comparative genomic hybridization
Abstract
Background: Small intestinal neuroendocrine tumors (SI-NETs) are typically slow-growing tumors that have metastasized already at the time of diagnosis. The purpose of the present study was to further refine and define regions of recurrent copy number (CN) alterations (CNA) in SI-NETs.
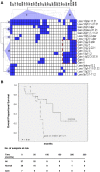
Methods: Genome-wide CNAs was determined by applying array CGH (a-CGH) on SI-NETs including 18 primary tumors and 12 metastases. Quantitative PCR analysis (qPCR) was used to confirm CNAs detected by a-CGH as well as to detect CNAs in an extended panel of SI-NETs. Unsupervised hierarchical clustering was used to detect tumor groups with similar patterns of chromosomal alterations based on recurrent regions of CN loss or gain. The log rank test was used to calculate overall survival. Mann-Whitney U test or Fisher's exact test were used to evaluate associations between tumor groups and recurrent CNAs or clinical parameters.
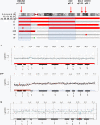
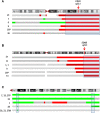
Results: The most frequent abnormality was loss of chromosome 18 observed in 70% of the cases. CN losses were also frequently found of chromosomes 11 (23%), 16 (20%), and 9 (20%), with regions of recurrent CN loss identified in 11q23.1-qter, 16q12.2-qter, 9pter-p13.2 and 9p13.1-11.2. Gains were most frequently detected in chromosomes 14 (43%), 20 (37%), 4 (27%), and 5 (23%) with recurrent regions of CN gain located to 14q11.2, 14q32.2-32.31, 20pter-p11.21, 20q11.1-11.21, 20q12-qter, 4 and 5. qPCR analysis confirmed most CNAs detected by a-CGH as well as revealed CNAs in an extended panel of SI-NETs. Unsupervised hierarchical clustering of recurrent regions of CNAs revealed two separate tumor groups and 5 chromosomal clusters. Loss of chromosomes 18, 16 and 11 and gain of chromosome 20 were found in both tumor groups. Tumor group II was enriched for alterations in chromosome cluster-d, including gain of chromosomes 4, 5, 7, 14 and gain of 20 in chromosome cluster-b. Gain in 20pter-p11.21 was associated with short survival. Statistically significant differences were observed between primary tumors and metastases for loss of 16q and gain of 7.
Conclusion: Our results revealed recurrent CNAs in several candidate regions with a potential role in SI-NET development. Distinct genetic alterations and pathways are involved in tumorigenesis of SI-NETs.
Figures
Similar articles
-
Genomic landscape of pancreatic neuroendocrine tumors.World J Gastroenterol. 2014 Dec 14;20(46):17498-506. doi: 10.3748/wjg.v20.i46.17498. World J Gastroenterol. 2014. PMID: 25516664 Free PMC article.
-
Global hypomethylation and promoter methylation in small intestinal neuroendocrine tumors: an in vivo and in vitro study.Epigenetics. 2014 Jul;9(7):987-97. doi: 10.4161/epi.28936. Epub 2014 Apr 24. Epigenetics. 2014. PMID: 24762809 Free PMC article.
-
Identification of novel genomic aberrations in AML-M5 in a level of array CGH.PLoS One. 2014 Apr 11;9(4):e87637. doi: 10.1371/journal.pone.0087637. eCollection 2014. PLoS One. 2014. PMID: 24727659 Free PMC article.
-
Genetics and epigenetics in small intestinal neuroendocrine tumours.J Intern Med. 2016 Dec;280(6):584-594. doi: 10.1111/joim.12526. Epub 2016 Jun 16. J Intern Med. 2016. PMID: 27306880 Review.
-
Recent advances in the molecular landscape of lung neuroendocrine tumors.Expert Rev Mol Diagn. 2019 Apr;19(4):281-297. doi: 10.1080/14737159.2019.1595593. Epub 2019 Mar 27. Expert Rev Mol Diagn. 2019. PMID: 30900485 Review.
Cited by
-
Genomic landscape of pancreatic neuroendocrine tumors.World J Gastroenterol. 2014 Dec 14;20(46):17498-506. doi: 10.3748/wjg.v20.i46.17498. World J Gastroenterol. 2014. PMID: 25516664 Free PMC article.
-
Clinical and Preclinical Advances in Gastroenteropancreatic Neuroendocrine Tumor Therapy.Front Endocrinol (Lausanne). 2017 Dec 4;8:341. doi: 10.3389/fendo.2017.00341. eCollection 2017. Front Endocrinol (Lausanne). 2017. PMID: 29255447 Free PMC article. Review.
-
Functional Copy-Number Alterations as Diagnostic and Prognostic Biomarkers in Neuroendocrine Tumors.Int J Mol Sci. 2024 Jul 9;25(14):7532. doi: 10.3390/ijms25147532. Int J Mol Sci. 2024. PMID: 39062773 Free PMC article.
-
Whole-genome sequencing of single circulating tumor cells from neuroendocrine neoplasms.Endocr Relat Cancer. 2021 Aug 11;28(9):631-644. doi: 10.1530/ERC-21-0179. Endocr Relat Cancer. 2021. PMID: 34280125 Free PMC article.
-
The Molecular Biology of Midgut Neuroendocrine Neoplasms.Endocr Rev. 2024 May 7;45(3):343-350. doi: 10.1210/endrev/bnad034. Endocr Rev. 2024. PMID: 38123518 Free PMC article. Review.
References
-
- Jann H, Roll S, Couvelard A, Hentic O, Pavel M, Muller-Nordhorn J, Koch M, Rocken C, Rindi G, Ruszniewski P. et al.Neuroendocrine tumors of midgut and hindgut origin: tumor-node-metastasis classification determines clinical outcome. Cancer. 2011;117(15):3332–3341. doi: 10.1002/cncr.25855. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

