A small-molecule inhibitor of hepatitis C virus infectivity
- PMID: 24165192
- PMCID: PMC3910743
- DOI: 10.1128/AAC.02083-13
A small-molecule inhibitor of hepatitis C virus infectivity
Abstract
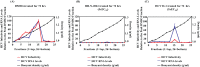
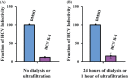
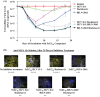
One of the most challenging goals of hepatitis C virus (HCV) research is to develop well-tolerated regimens with high cure rates across a variety of patient populations. Such a regimen will likely require a combination of at least two distinct direct-acting antivirals (DAAs). Combining two or more DAAs with different resistance profiles increases the number of mutations required for viral breakthrough. Currently, most DAAs inhibit HCV replication. We recently reported that the combination of two distinct classes of HCV inhibitors, entry inhibitors and replication inhibitors, prolonged reductions in extracellular HCV in persistently infected cells. We therefore sought to identify new inhibitors targeting aspects of the HCV replication cycle other than RNA replication. We report here the discovery of the first small-molecule HCV infectivity inhibitor, GS-563253, also called HCV infectivity inhibitor 1 (HCV II-1). HCV II-1 is a substituted tetrahydroquinoline that selectively inhibits genotype 1 and 2 HCVs with low-nanomolar 50% effective concentrations. It was identified through a high-throughput screen and subsequent chemical optimization. HCV II-1 only permits the production and release of noninfectious HCV particles from cells. Moreover, infectious HCV is rapidly inactivated in its presence. HCV II-1 resistance mutations map to HCV E2. In addition, HCV-II prevents HCV endosomal fusion, suggesting that it either locks the viral envelope in its prefusion state or promotes a viral envelope conformation change incapable of fusion. Importantly, the discovery of HCV II-1 opens up a new class of HCV inhibitors that prolong viral suppression by HCV replication inhibitors in persistently infected cell cultures.
Figures




Similar articles
-
Dehydrojuncusol, a Natural Phenanthrene Compound Extracted from Juncus maritimus, Is a New Inhibitor of Hepatitis C Virus RNA Replication.J Virol. 2019 May 1;93(10):e02009-18. doi: 10.1128/JVI.02009-18. Print 2019 May 15. J Virol. 2019. PMID: 30842319 Free PMC article.
-
Hepatitis C viral entry inhibitors prolong viral suppression by replication inhibitors in persistently-infected Huh7 cultures.PLoS One. 2013 Jun 3;8(6):e65273. doi: 10.1371/journal.pone.0065273. Print 2013. PLoS One. 2013. PMID: 23755208 Free PMC article.
-
Kinetic analyses reveal potent and early blockade of hepatitis C virus assembly by NS5A inhibitors.Gastroenterology. 2014 Aug;147(2):453-62.e7. doi: 10.1053/j.gastro.2014.04.021. Epub 2014 Apr 22. Gastroenterology. 2014. PMID: 24768676 Free PMC article.
-
Small molecule inhibitors of the hepatitis C virus-encoded NS5A protein.Virus Res. 2012 Dec;170(1-2):1-14. doi: 10.1016/j.virusres.2012.09.007. Epub 2012 Sep 23. Virus Res. 2012. PMID: 23009750 Review.
-
Antiviral activity and resistance of HCV NS5A replication complex inhibitors.Curr Opin Virol. 2013 Oct;3(5):514-20. doi: 10.1016/j.coviro.2013.06.014. Epub 2013 Jul 27. Curr Opin Virol. 2013. PMID: 23896281 Review.
Cited by
-
Identification of a resveratrol tetramer as a potent inhibitor of hepatitis C virus helicase.Br J Pharmacol. 2016 Jan;173(1):191-211. doi: 10.1111/bph.13358. Epub 2015 Nov 25. Br J Pharmacol. 2016. PMID: 26445091 Free PMC article.
-
How hepatitis C virus invades hepatocytes: the mystery of viral entry.World J Gastroenterol. 2014 Apr 7;20(13):3457-67. doi: 10.3748/wjg.v20.i13.3457. World J Gastroenterol. 2014. PMID: 24707128 Free PMC article. Review.
-
Entry Inhibitors of Hepatitis C Virus.Adv Exp Med Biol. 2022;1366:207-222. doi: 10.1007/978-981-16-8702-0_13. Adv Exp Med Biol. 2022. PMID: 35412143
-
Hepatitis C virus cell entry: a target for novel antiviral strategies to address limitations of direct acting antivirals.Hepatol Int. 2016 Sep;10(5):741-8. doi: 10.1007/s12072-016-9724-7. Epub 2016 Apr 5. Hepatol Int. 2016. PMID: 27048616 Free PMC article. Review.
-
Chlorcyclizine Inhibits Viral Fusion of Hepatitis C Virus Entry by Directly Targeting HCV Envelope Glycoprotein 1.Cell Chem Biol. 2020 Jul 16;27(7):780-792.e5. doi: 10.1016/j.chembiol.2020.04.006. Epub 2020 May 7. Cell Chem Biol. 2020. PMID: 32386595 Free PMC article.
References
-
- Sarrazin C, Kieffer TL, Bartels D, Hanzelka B, Muh U, Welker M, Wincheringer D, Zhou Y, Chu HM, Lin C, Weegink C, Reesink H, Zeuzem S, Kwong AD. 2007. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132:1767–1777. 10.1053/j.gastro.2007.02.037 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

