Resveratrol and EGCG bind directly and distinctively to miR-33a and miR-122 and modulate divergently their levels in hepatic cells
- PMID: 24165878
- PMCID: PMC3902894
- DOI: 10.1093/nar/gkt1011
Resveratrol and EGCG bind directly and distinctively to miR-33a and miR-122 and modulate divergently their levels in hepatic cells
Abstract
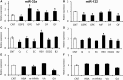
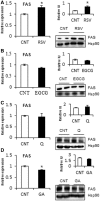
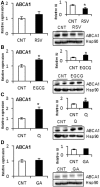
Modulation of miR-33 and miR-122 has been proposed to be a promising strategy to treat dyslipidemia and insulin resistance associated with obesity and metabolic syndrome. Interestingly, specific polyphenols reduce the levels of these mi(cro)RNAs. The aim of this study was to elucidate the effect of polyphenols of different chemical structure on miR-33a and miR-122 expression and to determine whether direct binding of the polyphenol to the mature microRNAs (miRNAs) is a plausible mechanism of modulation. The effect of two grape proanthocyanidin extracts, their fractions and pure polyphenol compounds on miRNA expression was evaluated using hepatic cell lines. Results demonstrated that the effect on miRNA expression depended on the polyphenol chemical structure. Moreover, miR-33a was repressed independently of its host-gene SREBP2. Therefore, the ability of resveratrol and epigallocatechin gallate to bind miR-33a and miR-122 was measured using (1)H NMR spectroscopy. Both compounds bound miR-33a and miR-122 and differently. Interestingly, the nature of the binding of these compounds to the miRNAs was consistent with their effects on cell miRNA levels. Therefore, the specific and direct binding of polyphenols to miRNAs emerges as a new posttranscriptional mechanism by which polyphenols could modulate metabolism.
Figures


References
-
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

