EBI metagenomics--a new resource for the analysis and archiving of metagenomic data
- PMID: 24165880
- PMCID: PMC3965009
- DOI: 10.1093/nar/gkt961
EBI metagenomics--a new resource for the analysis and archiving of metagenomic data
Abstract
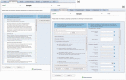
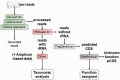
Metagenomics is a relatively recently established but rapidly expanding field that uses high-throughput next-generation sequencing technologies to characterize the microbial communities inhabiting different ecosystems (including oceans, lakes, soil, tundra, plants and body sites). Metagenomics brings with it a number of challenges, including the management, analysis, storage and sharing of data. In response to these challenges, we have developed a new metagenomics resource (http://www.ebi.ac.uk/metagenomics/) that allows users to easily submit raw nucleotide reads for functional and taxonomic analysis by a state-of-the-art pipeline, and have them automatically stored (together with descriptive, standards-compliant metadata) in the European Nucleotide Archive.
Figures

References
-
- Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 1998;5:R245–R249. - PubMed
-
- Hess M, Sczyrba A, Egan A, Kim T, Chokhawala H, Schroth G, Luo S, Clark DS, Chen F, Zhang T, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331:463–467. - PubMed
-
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. - PubMed
Publication types
MeSH terms
Grants and funding
- BB/H024921/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I02612X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I000771/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I025840/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E025080/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

