Distinct physiological and developmental properties of hippocampal CA2 subfield revealed by using anti-Purkinje cell protein 4 (PCP4) immunostaining
- PMID: 24166578
- PMCID: PMC4001794
- DOI: 10.1002/cne.23486
Distinct physiological and developmental properties of hippocampal CA2 subfield revealed by using anti-Purkinje cell protein 4 (PCP4) immunostaining
Abstract
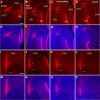
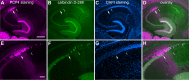
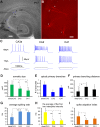
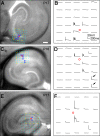
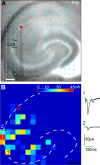
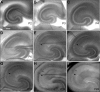
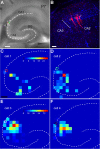
The hippocampal CA2 subfield was initially identified by Lorente de Nó as an anatomically distinct region based on its cytoarchitectural features. Although there is an enormous body of literature on other hippocampal subfields (CA1 and CA3), relatively little is known about the physiological and developmental properties of CA2. Here we report identification of the CA2 region in the mouse by immunostaining with a Purkinje cell protein 4 (PCP4) antibody, which effectively delineates CA3/CA2 and CA2/CA1 borders and agrees well with previous cytoarchitectural definitions of CA2. The PCP4 immunostaining-delineated CA2 neurons have distinguishable differences in cell morphology, physiology, and synaptic circuit connections compared with distal CA3 and proximal CA1 regions. The average somatic sizes of excitatory cells differ across CA1-3, with the smallest to largest somatic size being CA1<CA2<CA3. CA2 excitatory cells have dense dendritic spines, but do not have thorny excrescences associated with bordering CA3 neurons. Photostimulation functional circuit mapping shows that CA2 excitatory neurons receives extensive synaptic input from CA3, but no detectable input from the dentate gyrus. CA2 excitatory cells also differ significantly from CA3 cells in intrinsic electrophysiological parameters, such as membrane capacitance and spiking rates. Although CA2 neurons differ from CA1 neurons for PCP4 and other marker expressions, these neurons have less distinct neurophysiological and morphological properties. Developmental examination revealed that PCP4 immunostaining first appears at postnatal day 4-5 and becomes successively more refined around CA2 until reaching adult form by postnatal day 21.
Keywords: circuit mapping; electrophysiology; immunochemical.
Copyright © 2013 Wiley Periodicals, Inc.
Figures







References
-
- Bischofberger J, Engel D, Li L, Geiger JR, Jonas P. Patch-clamp recording from mossy fiber terminals in hippocampal slices. Nat Protoc. 2006;1:2075–2081. - PubMed
-
- Bland ST, Tamlyn JP, Barrientos RM, Greenwood BN, Watkins LR, Campeau S, Day HE, Maier SF. Expression of fibroblast growth factor-2 and brain-derived neurotrophic factor mRNA in the medial prefrontal cortex and hippocampus after uncontrollable or controllable stress. Neuroscience. 2007;144:1219–1228. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

