Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood
- PMID: 24166615
- PMCID: PMC3830740
- DOI: 10.1007/s00401-013-1199-1
Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood
Abstract
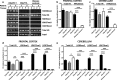
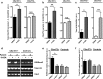
Individuals carrying (GGGGCC) expanded repeats in the C9orf72 gene represent a significant portion of patients suffering from amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Elucidating how these expanded repeats cause "c9FTD/ALS" has since become an important goal of the field. Toward this end, we sought to investigate whether epigenetic changes are responsible for the decrease in C9orf72 expression levels observed in c9FTD/ALS patients. We obtained brain tissue from ten c9FTD/ALS individuals, nine FTD/ALS cases without a C9orf72 repeat expansion, and nine disease control participants, and generated fibroblastoid cell lines from seven C9orf72 expanded repeat carriers and seven participants carrying normal alleles. Chromatin immunoprecipitation using antibodies for histone H3 and H4 trimethylated at lysines 9 (H3K9), 27 (H3K27), 79 (H3K79), and 20 (H4K20) revealed that these trimethylated residues bind strongly to C9orf72 expanded repeats in brain tissue, but not to non-pathogenic repeats. Our finding that C9orf72 mRNA levels are reduced in the frontal cortices and cerebella of c9FTD/ALS patients is consistent with trimethylation of these histone residues, an event known to repress gene expression. Moreover, treating repeat carrier-derived fibroblasts with 5-aza-2-deoxycytidine, a DNA and histone demethylating agent, not only decreased C9orf72 binding to trimethylated histone residues, but also increased C9orf72 mRNA expression. Our results provide compelling evidence that trimethylation of lysine residues within histones H3 and H4 is a novel mechanism involved in reducing C9orf72 mRNA expression in expanded repeat carriers. Of importance, we show that mutant C9orf72 binding to trimethylated H3K9 and H3K27 is detectable in blood of c9FTD/ALS patients. Confirming these exciting results using blood from a larger cohort of patients may establish this novel epigenetic event as a biomarker for c9FTD/ALS.
Figures



Comment in
-
Making sense of the antisense transcripts in C9FTD/ALS.Acta Neuropathol. 2013 Dec;126(6):785-7. doi: 10.1007/s00401-013-1201-y. Acta Neuropathol. 2013. PMID: 24178412 Free PMC article. No abstract available.
References
-
- Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78(2):547–581. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

